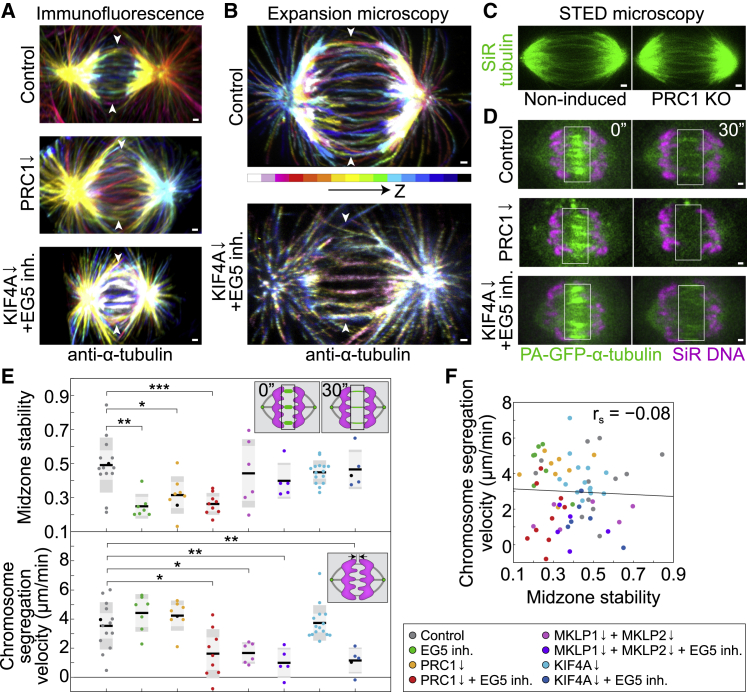
Figure 5.
Decreased midzone stability does not affect chromosome segregation velocity during anaphase
(A and B) Immunofluorescent (A) and expansion microscopy (B) images of fixed control, PRC1-siRNA-depleted (only immunofluorescence) and KIF4A-siRNA-depleted 40-μM STLC-treated RPE-1 cells stained with AlexaFluor594 conjugated with α-tubulin antibody. Images in (A) and (B) are the maximum projections of color-coded z stacks as shown by the scheme in (B). Arrowheads point to the spindle midzone region.
(C) STED images (single z plane) of anaphase spindles labeled with 100 nM SiR-tubulin in live non-induced CRISPR PRC1 KO and induced PRC1 KO RPE-1 cells.
(D) Smoothed live-cell images (single z plane) of RPE-1 cells after midzone photoactivation of photoactivatable (PA)-GFP-α-tubulin (green) in the indicated conditions. 100-nM SiR-DNA (magenta) was used for chromosome staining. Time 0 represents the photoactivation onset.
(E) Quantification of midzone stability, defined as the ratio of the integrated photoactivated signal in the boxed region (see schemes and boxes in D) at times 30 and 0 s, and chromosome segregation velocity measured in the same time period (see schemes), for the indicated treatments. Statistics: t test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001). Numbers: 14, 7, 8, 9, 6, 5, 15, and 5 cells, from left to right. Three independent experiments for every condition except MKLP1 + MKLP2, MKLP1 + MKLP2 + EG5 inh. and KIF4+EG5 inh., which were done in two independent experiments regarding siRNAs or non-targeting treatments, while number of independent experiments regarding STLC treatment is equal to the number of cells. Black data dots in every treatment correspond to the measurements from the exemplar cells shown on the time-lapse images and kymographs (D).
(F) Chromosome segregation velocity versus midzone stability (data from E) for indicated treatments and linear regression (line); rs, Spearman correlation coefficient, p < 0.001. Scale bars, 1 μm. See also Figure S5.