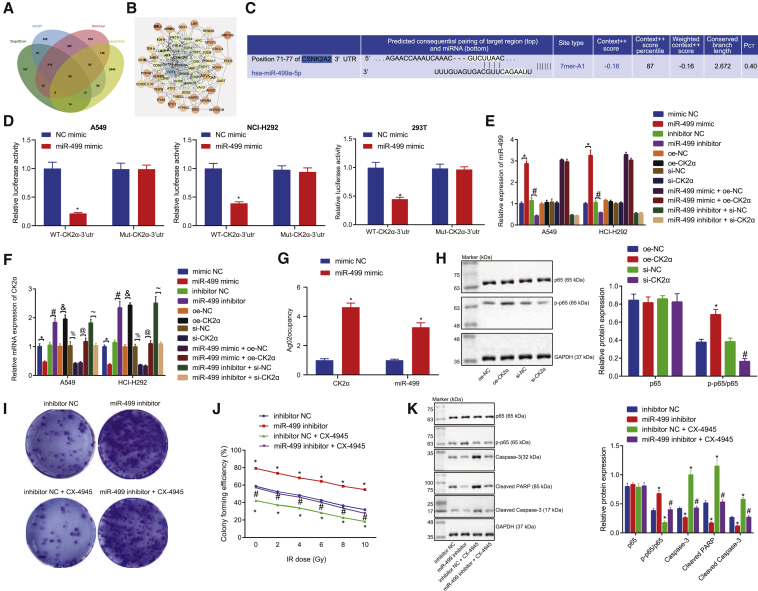
Figure 5.
miR-499 promoted the sensitivity of lung cancer cells to IR exposure by targeting CK2α and blocking p65 phosphorylation
(A) Venn diagram displaying the intersection of downstream target gene results of miR-499 predicted by TargetScan, mirDIP, and RNAInter bioinformatics databases and NSCLC-related genes in GeneCards database. (B) STRING was used to analyze the network diagram of interaction relationships between candidate genes, with circles ranging in color from orange to blue indicating the increasing degree of genes and lines in the middle of circles indicating interaction relationships between genes. (C) Prediction of relationship between miR-499 and CK2α by a bioinformatics website. (D) Dual-luciferase activity in HEK293T cells detected by dual-luciferase gene assay. (E and F) The regulatory relationship between miR-499 and CK2α verified by qRT-PCR. ∗p < 0.05 versus cells treated with NC mimic; #p < 0.05 versus cells transfected with inhibitor NC (E and F); &p < 0.05 versus upon treatment of oe-NC; %p < 0.05 versus upon treatment of si-NC; @p < 0.05 versus cells treated with miR-499 mimic and oe-NC; ~p < 0.05 versus cells transfected with miR-499 inhibitor and si-NC. (G) RIP assay verifying the binding of miR-499 to CK2α. (H) The regulatory relationship between CK2α and p65 phosphorylation verified by western blot analysis in A549 cells. (I) Sensitivity of lung cancer cells to radiation as determined by MTT assay. ∗p < 0.05 versus cells treated with oe-NC; #p < 0.05 versus cells transfected with si-NC. (J) The effect of miR-499 through targeting CK2α on the sensitivity of radiotherapy after 10 Gy IR verified by colony-formation assay in A549 cells. ∗p < 0.05 versus cells treated with miR-499 inhibitor; #p < 0.05 versus cells treated with inhibitor NC + CX-4945. (K) The effects of miR-499 on p65, phosphorylated p65/p65, cleaved PARP, caspase-3, and cleaved caspase-3 expression determined by western blot analysis in A549 cells. ∗p < 0.05 versus cells treated with miR-499 inhibitor; #p < 0.05 versus cells treated with inhibitor NC + CX-4945. The measurement data were expressed as mean ± standard deviation. Data between two groups were compared using independent sample t test.