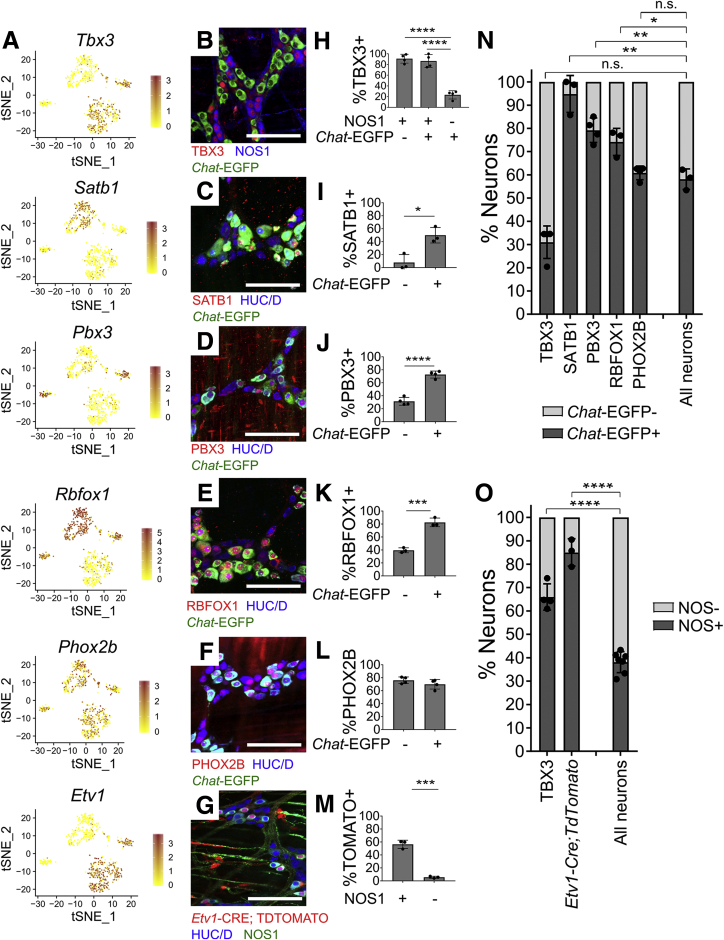
Figure 8.
Regulatory genes are expressed in distinct adult ENS subsets. (A) Feature plots of select regulatory genes. Color key shows loge(normalized gene expression). (B–F) Myenteric plexus wholemount immunohistochemistry in young adult ChAT-EGFP-L10A distal colon. (G) Whole mount myenteric plexus immunohistochemistry in distal colon from tamoxifen-treated Etv1-CreErt2;R26R-TdTomato mice. TDTOMATO is in many NOS1+ neurons and some non-neuronal cells with ICC morphology. (H–L) Immunohistochemistry quantification. TBX3 is found mostly in (H) nitrergic (NOS1+) neurons (P < .0001, n = 3). (I) SATB1 (P = .013, n = 3), (J) PBX3 (P < .0001, n = 3), and (K) RBFOX1 (P = .0006, n = 3) are mostly in cholinergic (EGFP+) neurons, consistent with single-cell RNA data. (L) PHOX2B is equally abundant in Chat-EGFP+ and Chat-EGFP–cells (P = .2193, n = 4). (M) Quantification of G shows that 56.4 ± 3.6% of NOS1+ neurons are TDTOMATO+. Only 5.6 ± 0.9% of NOS1– neurons are TDTOMATO+ (P = .0002, n = 3). (N) Cholinergic (Chat-EGFP+) and (O) nitrergic (NOS1+) identity for cells expressing select factors. SATB1 (P = .0022), PBX3 (P = .0026), and RBFOX1 (P = .019) are largely restricted to cholinergic (Chat-GFP+) neurons. PHOX2B is present in both cholinergic and noncholinergic neurons (P = .370) (P values compare Transcription factor+EGFP+/Transcription factor+ vs EGFP+/Total neuron ratios). TBX3 (P < .0001) and ETV1 (P < .0001) are primarily in nitrergic (NOS1+) neurons (P values compare Transcription factor+NOS1+/Transcription factor+ vs NOS1+/Total neuron ratios). (I–O) Student’s t test. (H) Analysis of variance with Tukey’s post hoc test. Scale bar = 100 μm. ChAT-EGFP-L10A = Chat-EGFP. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001.