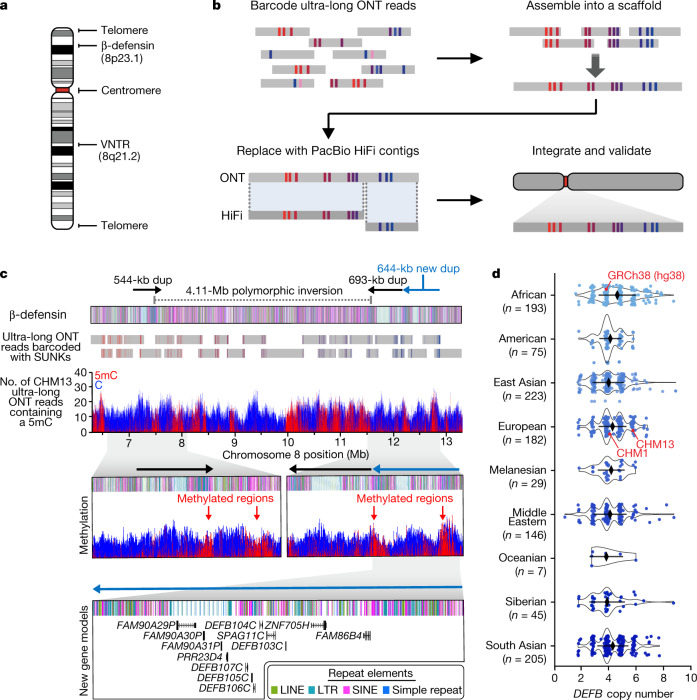
Fig. 1. Telomere-to-telomere assembly of human chromosome 8.
a, Gaps in the GRCh38 chromosome 8 reference sequence. b, Targeted assembly method to resolve complex repeat regions in the human genome. Ultra-long ONT reads (grey) are barcoded with SUNKs (coloured bars) and assembled into a sequence scaffold. Regions within the scaffold sharing high sequence identity with PacBio HiFi contigs (dark grey) are replaced, improving the base accuracy to greater than 99.99%. The PacBio HiFi assembly is integrated into an assembly of CHM13 chromosome 8 (ref. 5) and validated. c, Sequence, structure, methylation status and genetic composition of the CHM13 β-defensin locus. The locus contains three segmental duplications (dups) at chr8:7098892–7643091, chr8:11528114–12220905 and chr8:12233870–12878079. A 4,110,038-bp inversion (chr8:7500325–11610363) separates the first and second duplications. Iso-Seq data reveal that the third duplication (light blue) contains 12 new protein-coding genes, five of which are DEFB genes (Extended Data Fig. 3g). d, Copy number of the DEFB genes (chr8:7783837−7929198 in GRCh38) throughout the human population, determined from a collection of 1,105 high-coverage genomes (Methods). Data are median ± s.d.