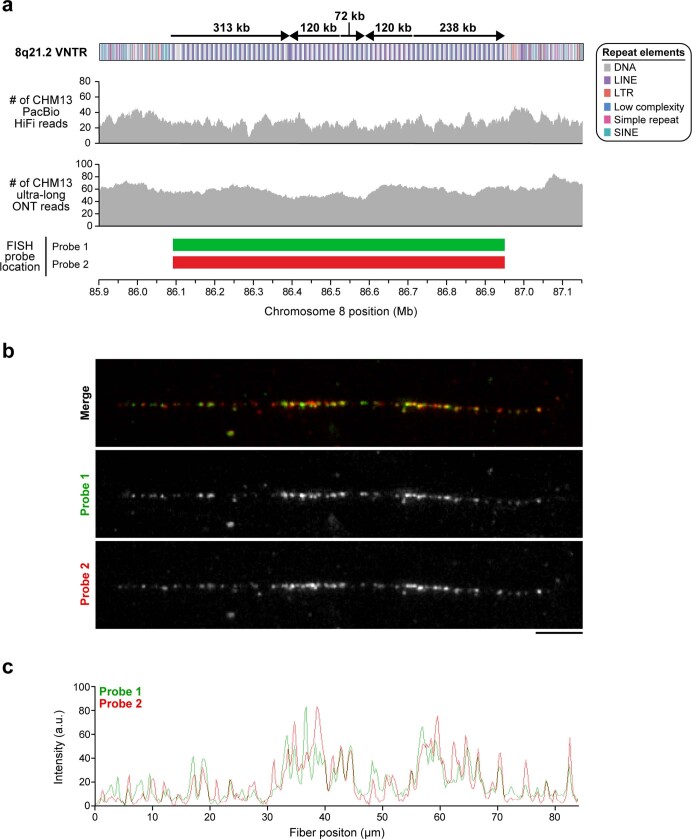
Extended Data Fig. 10. Validation of the CHM13 8q21.2 VNTR.
a, Coverage of CHM13 ONT and PacBio HiFi data along the 8q21.2 VNTR (top two panels) is largely uniform, indicating a lack of large structural errors. Two FISH probes targeting the 12.192-kb repeat in the 8q21.2 VNTR are used to estimate the number of repeats in the CHM13 genome (b, c). b, Representative FISH images of a CHM13 stretched chromatin fibre. Although the FISH probes were designed against the entire VNTR array, stringent washing during FISH produces a punctate probe signal pattern, which may be due to stronger hybridization of the probe to a specific region in the 12.192-kb repeat (perhaps based on GC content or a lack of secondary structures). This punctate pattern can be used to estimate the repeat copy number in the VNTR, thereby serving as a source of validation. c, Plot of the signal intensity on the CHM13 chromatin fibre shown in b. Quantification of peaks across three independent experiments reveals an average of 63 ± 7.55 peaks and 67 ± 5.20 peaks (mean ± s.d.) from the green and red probes, respectively, which is consistent with the number of repeat units in the 8q21.2 assembly (67 full and 7 partial repeats). Scale bar, 5 μm.