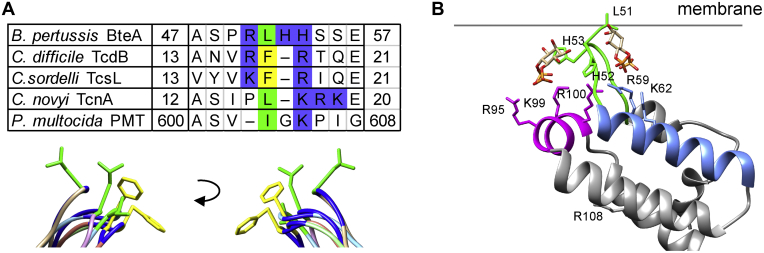
Figure 7.
Comparison of LRT with related PDB structures and its membrane association model.A, amino acid and structure alignments of the L1 region. Amino acids I/L are highlighted in green, F are highlighted in yellow, H/K/R are highlighted in blue. The structural alignment of the LRT domain of B. pertussis BteA (brown, PDB code: 6RGN) with 4HBM domains of C. difficile TcdB (orange, PDB code: 5UQM), C. sordelii TcsL (light green, PDB code: 2VKD), C. novyi TcnA (pink, PDB code: 2VK9), and P. multocida PMT (light blue, PDB code: 2EBF) was performed using Chimera 1.14rc. B, model of membrane association of the LRT domain. Critical amino acid residues identified by domain mutagenesis are depicted as sticks within the ribbon representation of the LRT structure and were visualized by Chimera 1.14rc. The headgroup of PIP2 (inositol 4,5-bisphosphate) was docked by AutoDock Vina. The binding energy was almost equal for both ligand binding sites, exhibiting the binding affinity value of −3.7 kcal/mol. LRT, lipid raft targeting.