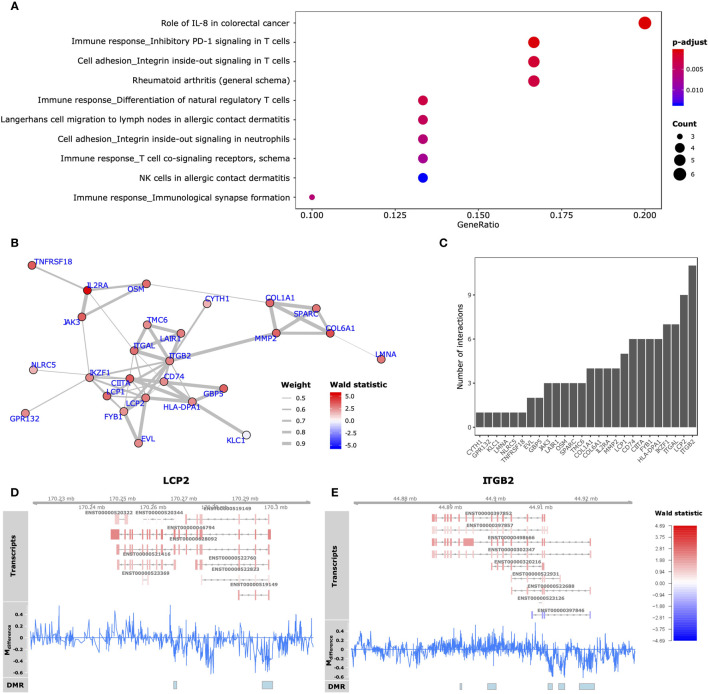
Figure 5.
Functional analyses GDEMs. (A) The 10 most overrepresented MetaCore gene sets found for the GDEMs depicting the ratio of pathway-associated GDEMs relative to the total number of pathway-associated genes. The size and colour intensity are proportional to the number of pathway-associated GDEMs and the -log10(adjusted p-values), respectively. (B) GDEMs were analysed for known interactions by querying the STRING database. Represented here is the protein-protein interaction network, where the thickness of the edge is proportional to the STRINGdb evidence score and the colour proportional to the Wald statistic, respectively. (C) Bargraph representing the number of interactions per protein. Summary visualization of the DNA methylation and gene expression signals for GDEMs: (D) LCP2, (E) ITGB2. The “Transcripts” track represents individual transcripts coloured by the Wald statistic. The “Mdifference” track represents the difference in percentage methylation when comparing SJC66high with SJC66low. The “DMR” track represents the locations of the DMRs as found by dmrseq.