Fig. 6.
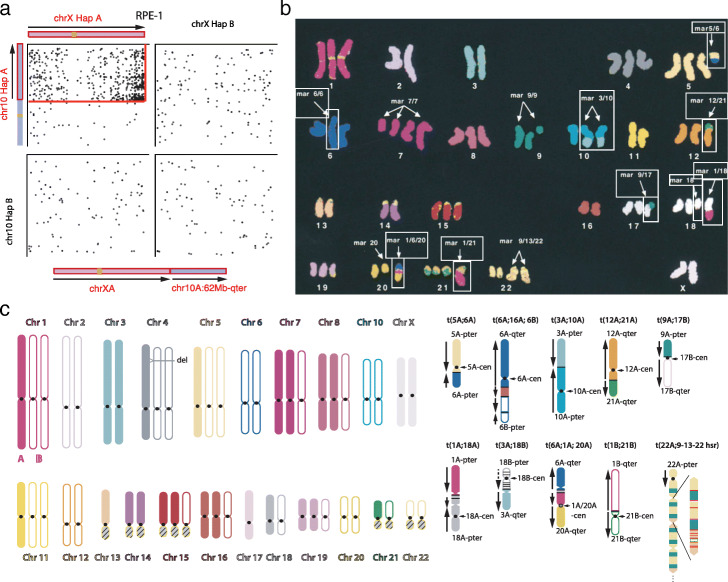
Haplotype-resolved synteny of rearranged chromosomes and aneuploid genomes. a Walking the translocated X chromosome in the RPE-1 genome using phased Hi-C links (dots) between different homologs of Chr.10 and Chr.X. A significant increase of Hi-C links is seen in only one haplotype combination reflecting cis links generated by the translocation between Chr.10 and Chr.X. The enrichment of Hi-C links throughout the entire X chromosome suggests the 10q segment is attached to an intact X chromosome with structure shown below the Hi-C contact map. Arrows denote the orientation of the two segments from the p-terminus to the q-terminus. b The cytogenetic K-562 karyotype reported in Ref. [42] (reprinted with permission from the publisher) with outlined structurally altered (marker) chromosomes resolved by sequencing data (shown in c). c The digital K-562 karyotype with haplotype assignment to both normal (left) and structurally altered (right) chromosomes determined from linked-reads and Hi-C sequencing data. The digital karyotype mostly agrees with the cytogenetic karyotype and the differences may be attributed to additional alterations during cell culture. Among all marker chromosomes listed in b, we are able to determine the syntenic structure of the following: mar5/6, mar6/6, mar3/10, mar12/21, mar9/17, mar1/18, mar1/6/20, and mar1/21, and resolve most rearrangement junctions at the base-pair level. Arrows represent the orientations of rearranged segments relative to the standard p-q arm orientation. Multiple junctions contained local fold-back rearrangements (inverted colored arrows in t(1A;18A), t(3A;18B), t(6A;1A;20A)) that are consistent with local DNA copy number gains; these events cannot be resolved by cytogenetic analyses. The mar18 described in Ref. [42] is probably related/similar to t(3A;18B). The BCR-ABL amplification is contained in a homogeneously staining region (hsr) in the marker chromosome t(22A;9-13-22hsr). We infer the structure of the amplicon from DNA copy number and rearrangements but cannot validate the inferred structure due to technical limitations. We are further able to partially resolve the structure of the altered Chr.7 and Chr.9 and completely resolve the structure of three additional marker chromosomes described in Ref. [43] but not in Ref. [42]: t(2A;22A), t(3A;10A;17A), t(9A;13A). Details of the analysis are presented in Additional file 5 and explained in Additional file 1:Determination of the K-562 karyotype by haplotype-specific genomic analysis