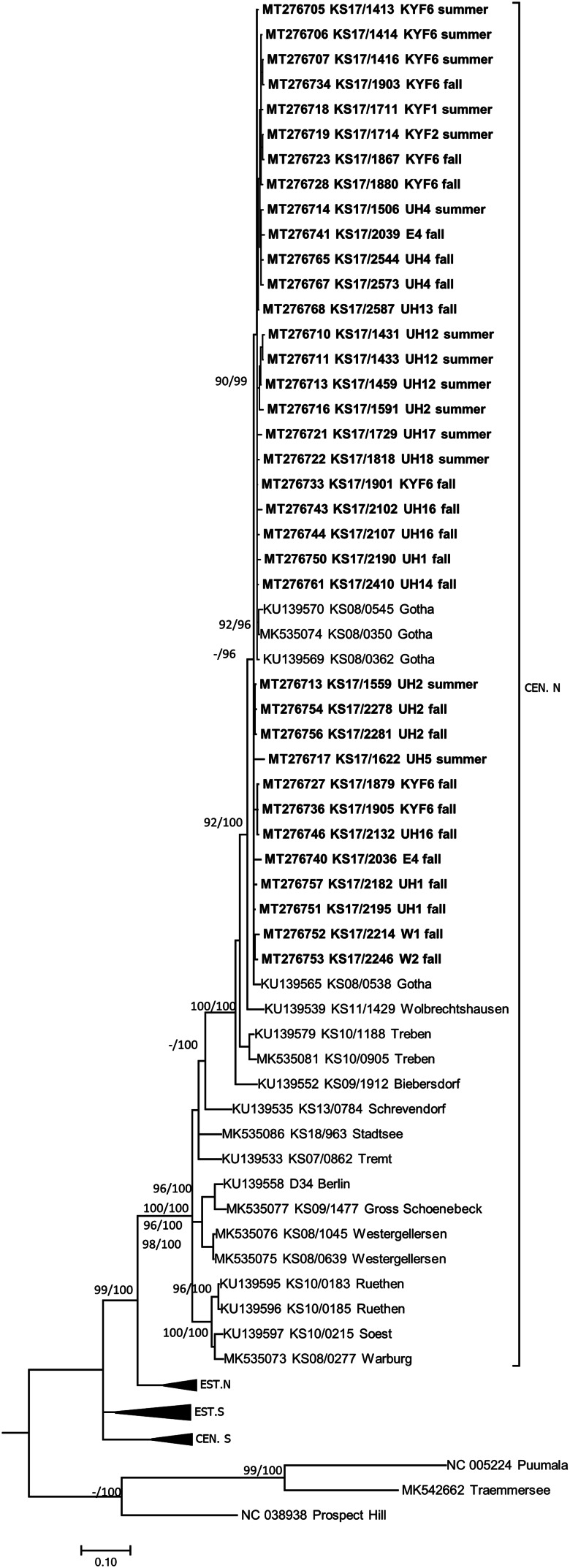
Fig. 2.
Consensus phylogenetic tree of the partial S segment sequences of Tula orthohantavirus (TULV) (alignment length 549 nucleotides (nt), positions 406–951, counting according to TULV S segment, accession number NC_005227). TULV is sorted in the clades Central North (CEN.N), Central South (CEN.S), Eastern North (EST.N) and Eastern South (EST.S). The consensus tree is based on Bayesian analyses with 107 generations and a burn-in phase of 25%, and maximum-likelihood analyses, with 1000 bootstraps and 50% cut-off using the general time reversible (GTR) substitution model with invariant sites and a gamma distributed shape parameter for both algorithms. Posterior probabilities exceeding 95% from Bayesian analyses are given behind and bootstrap values are given before the slash for major nodes if exceeding 75%. The tree reconstructions were done via CIPRES [23]. Alignments were constructed with Bioedit V7.2.3. [21] using the Clustal W Multiple Alignment algorithm implemented in the program. Names in bold indicate newly generated sequences (see Supplementary Table S1). Triangles indicate compressed branches (see Supplementary Table S2 for used sequences). Clade designation followed previous publications for TULV [28].