Figure 2.
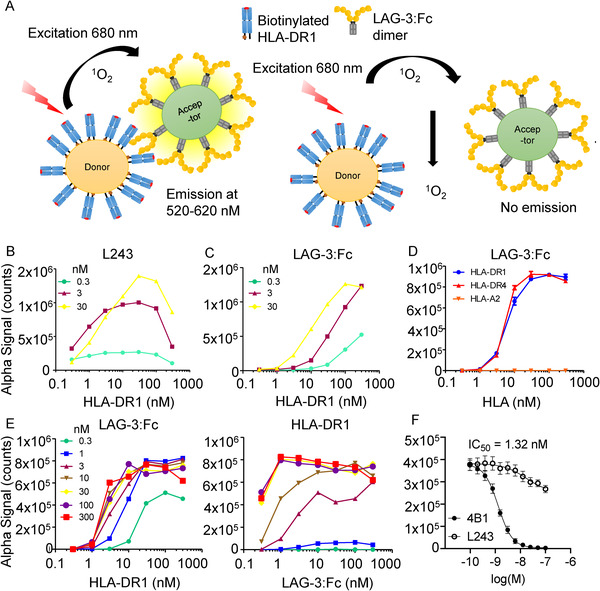
(A) Schematic overview of experimental setup for AlphaScreenTM assays. (B) L243 binding to pHLA‐DR1. Assays used three concentrations of L243 antibody (0.3, 3, and 30 nM), titrated against a one in three dilution series (0.3 to 300 nM) of pHLA‐DR1. Data are representative of three independent experimental repeats using freshly prepared beads. (C) LAG‐3:Fc binding to pHLA‐DR1. Assays used three concentrations of LAG‐3:Fc (0.3, 3, and 30 nM), titrated against a one in three dilution series (0.3 to 300 nM) of pHLA‐DR1. Data are representative of three independent experimental repeats using freshly prepared beads. (D) LAG‐3:Fc binding to pHLA‐DR1 and ‐DR4. Titration of pHLA‐DR1 and ‐DR4 in one in three dilution series (300 nM to 0.3 nM). Titration of biotinylated HLA‐A*02:01 (complexed with the hTERT540‐548 peptide) in a one in three dilution series (300 nM to 0.3 nM) was used as a negative control. Data are representative of three experimental repeats using freshly prepared beads. (E) Cross titrations using LAG‐3:Fc and pHLA‐DR1 using a one in three dilution series (300 nM to 0.3 nM). Data are representative of three biological repeats. (F) AlphaScreenTM LAG‐3:Fc/pHLA‐DR1 blockade assay using anti‐LAG‐3 4B1 and anti‐DR L243 fab fragments, IC50 = 1.32 nM. Data are inclusive of three independent experimental repeats using freshly prepared beads, error bars represent mean ± SEM.
