Figure 7.
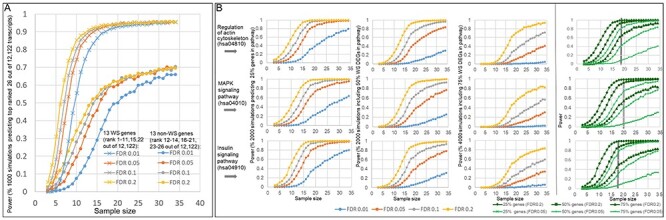
Simulated power curves of sample size. (A) Simulated power curves for detecting differential expression from smaller subsets of the data show that detection of the 13 WS genes (rank 1–11, 15, 22 out of 12 122 transcripts) approaches 90% power at ~9 samples, while detection of the top 13 non-WS genes (rank 12–14, 16–21, 23–26 out of 12 122 transcripts) approaches 60% power at ~20 samples (FDR = 0.2). (B) Simulated power curves for detecting differential expression from smaller subsets of the data show the sample size of detection of 25, 50 or 75% of the WS DEGs (FDR < 0.2) (Supplementary Material, Table S10) picked up by KEGG 2016 pathways (regulation of actin cytoskeleton (hsa04810); MAPK signaling pathway (hsa04010); insulin signaling pathway (hsa04910)). The simulations indicate that to identify the actin signaling pathway, when FDR = 0.2, at least 15 cases of the current dataset would have been needed to have detected at least 25% of the genes with 90% power, and to have detected 50% of the genes with 90% power, at least 20 cases would have been needed. For the insulin signaling pathway, 18 and 25 cases would have been needed to attain this power, and for the MAPK signaling pathway, 16 and 25.
