Abstract
Yeast decode pheromone gradients to locate mating partners, providing a model for chemotropism. How yeast polarize toward a single partner in crowded environments is unclear. Initially, cells often polarize in unproductive directions, but then they relocate the polarity site until two partners’ polarity sites align, whereupon the cells “commit” to each other by stabilizing polarity to promote fusion. Here we address the role of the early mobile polarity sites. We found that commitment by either partner failed if just one partner was defective in generating, orienting, or stabilizing its mobile polarity sites. Mobile polarity sites were enriched for pheromone receptors and G proteins, and we suggest that such sites engage in an exploratory search of the local pheromone landscape, stabilizing only when they detect elevated pheromone levels. Mobile polarity sites were also enriched for pheromone secretion factors, and simulations suggest that only focal secretion at polarity sites would produce high pheromone concentrations at the partner’s polarity site, triggering commitment.
INTRODUCTION
Directed growth (chemotropism) or movement (chemotaxis) in response to a chemical signal is critical for biological processes including aggregation in Dictyostelium discoideum (Nichols et al., 2015), pollen tube growth during plant fertilization (Higashiyama and Takeuchi, 2015), axon guidance during neural development (Bellon and Mann, 2018), and neutrophil migration in the mammalian immune response (Sarris and Sixt, 2015). However, the mechanisms by which cells choose the direction of polarized growth or movement are incompletely understood.
Yeast cells polarize secretion, and hence growth, toward their partners during mating (Merlini et al., 2013). Saccharomyces cerevisiae cells of each mating type, a and α, secrete pheromones that are sensed by cognate G protein–coupled receptors on cells of the opposite mating type. Pheromone sensing triggers the activation of a MAPK cascade, cell-cycle arrest in G1, increased transcription of mating-specific genes, and polarized growth toward the mating partner (Dohlman and Thorner, 2001).
Polarity is directed by the Rho-GTPase Cdc42, which becomes concentrated together with its regulators and effectors at a small region of the cell’s cortex to form a “polarity site” (Park and Bi, 2007). Polarity sites are assembled by a positive feedback mechanism in which active Cdc42-GTP binds the scaffold protein Bem1, promoting the activation of nearby inactive Cdc42-GDP to form a cluster of polarity factors (Kozubowski et al., 2008; Johnson et al., 2011). Formins (effectors of Cdc42) trigger the orientation of actin cables toward the site, promoting delivery of secretory vesicles (Pruyne et al., 2004).
Yeast cells exposed to a stable pheromone gradient tend to grow up-gradient (Segall, 1993), and stable gradients are often assumed to mediate partner selection (Figure 1A; Arkowitz, 1999; Ismael and Stone, 2017). However, stable pheromone gradients may be rare in the wild, where cells mate in the context of tetrads (Figure 1B; Taxis et al., 2005) or microcolonies (Figure 1C; McClure et al., 2018). Even when surrounded by several potential partners, cells choose only one (Jackson and Hartwell, 1990a,b). It is unclear how such mating geometries could produce stable pheromone gradients yielding orientation toward just one partner (Jin et al., 2011; Rappaport and Barkai, 2012).
FIGURE 1:
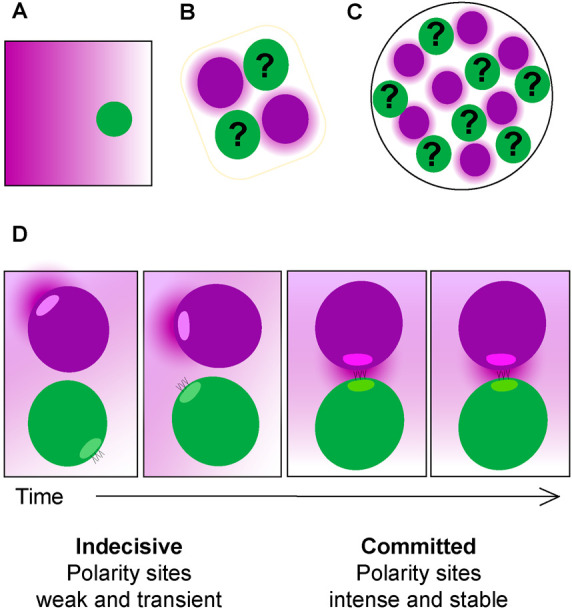
Pheromone landscapes encountered by yeast cells. (A) Stable unidirectional pheromone gradient, as generated by micropipette or microfluidics device. (B) Germinating spores in an ascus, where two potential partners (magenta) are expected to generate similar α-factor gradients, making them equally attractive to the a cells (green). (C) Microcolony containing a mixture of a (green) and α cells (magenta). The proximity of multiple potential partners complicates the task of orienting toward a single partner. (D) Exploratory polarization model of partner selection. During the indecisive period (frames 1 and 2), diffusion of pheromone released at the α cell’s (magenta) polarity site yields a low pheromone concentration at the a cell’s (green) polarity site. When the two polarity sites are apposed (frames 3 and 4), the a cell senses a high concentration of pheromone. Both cells sense and secrete pheromone, but for simplicity, only the a cell’s receptors and α cell’s pheromone are shown.
For successful mating, the pheromone landscape must be decoded to orient polarity toward the partner. However, imaging of polarity factors revealed that initial polarity sites were not always oriented toward the eventual mating partner (Henderson et al., 2019; Wang et al., 2019) or up-gradient in artificial pheromone gradients (Jin et al., 2011; Dyer et al., 2013; Hegemann et al., 2015; Kelley et al., 2015; Vasen et al., 2020). Rather, the location of the polarity site changed over time, stabilizing at better-oriented locations. In mating mixes, weak clusters of polarity factors appeared, disappeared, and changed position in a chaotic manner (Henderson et al., 2019). After this “indecisive phase” of 10–120 min, cells developed strong, stable polarity sites oriented toward the partner, suggesting that they made a “commitment” to the partner. Similarly, cells exposed to a steep pheromone gradient in a microfluidics device spent a variable interval with weak and mobile polarity sites before developing a strong polarity site at a stable position (Hegemann et al., 2015). These studies suggested that yeast cells process spatial information about the local pheromone landscape during a search period of variable duration, then commit to a specific orientation for polarized growth.
Spatial information about the pheromone landscape could be extracted by “global” or “local” sensing strategies (Kelley et al., 2015; Hegemann and Peter, 2017; Martin, 2019). In global sensing, cells compare the concentration of ligand-bound receptors around the cell surface to infer the direction of the pheromone source. In local sensing, cells primarily detect pheromone in a sensitized zone centered around the polarity site, moving the site around to infer the direction of the pheromone source. These models are not mutually exclusive.
Evidence for global sensing came from the observation that in mating mixes, initial weak polarity sites were oriented toward their eventual mating partners more often than would be expected by chance (Henderson et al., 2019). Thus, some spatial information was available before any polarity sites were visible. In principle, continued global sensing during the indecisive phase (when polarity sites are weak and mobile) could promote the selection of optimal locations for stable “committed” polarity sites.
Evidence for local sensing came from the observation that when a strong polarity site is present, pheromone receptors and associated G proteins accumulate around the polarity site (Ayscough and Drubin, 1998; Suchkov et al., 2010; McClure et al., 2015). Thus, cells with a strong polarity site sense pheromone preferentially in the vicinity of the site. It is unclear whether the weak and transient polarity sites characteristic of the indecisive phase would similarly enable local sensing.
In Schizosaccharomyces pombe, weak and transient polarity sites in mating cells are enriched for pheromone secretion factors, as well as pheromone sensing factors (Merlini et al., 2016). If such enrichment is functionally important, then mobile polarity sites might represent the predominant sites of pheromone emission. These and other observations suggested a potential strategy for partner search that we call “exploratory polarization” (Figure 1D; Henderson et al., 2019).
If cells secrete pheromone primarily from the indecisive polarity sites, then the pheromone landscape would change as the polarity sites move. If cells also sense pheromone primarily at the indecisive polarity sites, then the pheromone concentration they detect would depend on the distance to the nearest partner’s polarity site. When polarity sites are not properly aligned, the pheromone released from one site is dissipated by diffusion before it is detected at the partner’s polarity site (Figure 1D). However, when two polarity sites face each other (and only in that case), a high concentration of pheromone is detected at each site (Figure 1D). In both S. cerevisiae and S. pombe, detection of a high pheromone concentration stabilizes the location of the polarity site (Bendezu and Martin, 2013; Dyer et al., 2013; McClure et al., 2015; Merlini et al., 2016). Thus, coincident detection of high pheromone levels upon alignment of partner polarity sites could stabilize both sites, leading to coordinated commitment by both partners.
Although the exploratory polarization hypothesis suggests an appealing strategy for partner selection, it is unclear to what degree pheromone secretion actually becomes polarized during the indecisive phase. If most pheromone is emitted globally from the entire cell surface, then that would create a stable pheromone gradient that could be decoded via either global sensing (Henderson et al., 2019) or local sensing (Hegemann et al., 2015; Wang et al., 2019). In this study, we show that as in S. pombe, transient polarity sites in S. cerevisiae are often enriched in pheromone sensing, signaling, and secretion proteins. Computational simulations provide quantitative support for the idea that pheromone levels sufficient to promote commitment are probably only achieved when polarity sites of mating partners become aligned. Most critically, we show that wild-type cells are unable to commit to partners that are impaired in the formation, localization, or stabilization of indecisive phase polarity sites. We conclude that local sensing and secretion by two cooriented polarity sites enables commitment by a mating pair.
RESULTS
Pheromone sensing and secretion are enriched at transient polarity sites
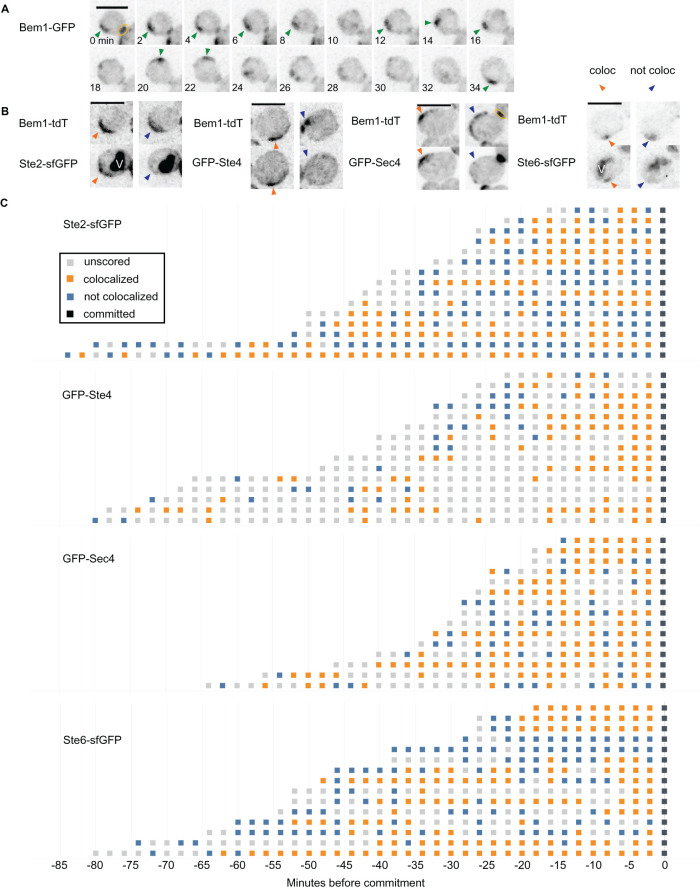
Pheromone receptors and G proteins are concentrated near stable polarity sites (Ayscough and Drubin, 1998; Suchkov et al., 2010; McClure et al., 2015), but it was unclear whether the transient polarity sites characteristic of the indecisive period would similarly concentrate these factors. We imaged strains harboring both BEM1-tdTomato to label the polarity site and either STE2-sfGFP (receptor) or GFP-STE4 (Gβ). In mating mixes, we identified time points at which Bem1 sites were clearly identifiable (Figure 2A) and assessed by visual comparison whether the Ste2 or Ste4 signal was colocalized with Bem1 (Figure 2B). Both the receptor and Gβ were sometimes, though not always, colocalized with polarity factors (Figures 2C and Supplemental Figure S1A).
FIGURE 2:
Localization of pheromone secretion, sensing, and signaling proteins during the indecisive period. (A) Example cell showing Bem1 (polarity marker) behavior during the indecisive phase. Strains harboring Bem1-tdTomato (DLY12943) were mixed with wild-type (DLY9070) and imaged. Scale bar: 4 µm. For scoring colocalization, Bem1 distributions were considered either scorable (one or two predominant Bem1 clusters or a Bem1 crescent; green arrowheads) or not scorable (diffuse, weak, and/or multiple Bem1 clusters; no arrowhead). See Supplemental Figure S1. (B) Example images from scorable time points during the indecisive phase, in which the indicated probes were scored as colocalized (orange arrowhead) or not colocalized (blue arrowhead) with Bem1. Strains harboring Bem1-tdTomato and either the α-factor receptor Ste2-sfGFP (DLY22243), Gβ subunit GFP-Ste4 (DLY23354), secretory vesicle marker GFP-Sec4 (DLY13771), or a-factor transporter Ste6-sfGFP (DLY22355) were mixed with wild-type (DLY8156) and imaged. Internal signal in Ste2-sfGFP and Ste6-sfGFP strains is due to sfGFP accumulation in the vacuole (V) following Ste2/Ste6 degradation. Yellow oval: cytokinesis site. Scale bars: 5 µm. (C) Cell behaviors during the indecisive phase, scored from the same mating mixes as in B. Cells that experienced an indecisive period of more than five time points were tracked either from the beginning of the movie (if already in G1) or from birth (appearance of Bem1 at the neck) until commitment. Each row represents one cell: each time point was designated as not scorable (gray box) or scorable, and the latter were scored as colocalized (orange box) or not colocalized (blue box). x-Axis: minutes before commitment.
We also examined pheromone secretion factors. The two pheromones are secreted by different mechanisms: α-factor is delivered to the plasma membrane in secretory vesicles, and a-factor is secreted by a transporter, Ste6 (Michaelis and Barrowman, 2012). As with the pheromone sensing probes, GFP-Sec4 (a marker of secretory vesicles) and Ste6-sfGFP were often enriched at indecisive polarity sites (Figure 2B and Supplemental Figure S1A). Because colocalization can occur while Bem1-containing polarity sites are still mobile, these data suggest that even transient polarity sites can create detectable enrichment of pheromone sensing and secretion proteins.
Indecisive polarity site behavior was also seen in cells treated with a low but uniform level of α-factor (Supplemental Figure S1B), suggesting that such behavior reflects the overall pheromone sensed by the cell and does not require a pheromone gradient. Nevertheless, the frequent orientation of polarity sites toward potential partners during the indecisive stage (Henderson et al., 2019) suggests that polarity location is influenced by pheromone gradients at this stage.
Simulating the pheromone landscape experienced by mating cells
Our findings support the possibility that pheromone is emitted locally from the polarity site. To understand how local, as opposed to global, pheromone emission would affect the pheromone landscape sensed by the partner, we first used solutions of the diffusion equation to estimate the pheromone concentration that is expected to be detected next to a cell secreting pheromone. An α cell exposed to a-factor secretes approximately 1400 molecules of α-factor per second (Rogers et al., 2012). Assuming that the pheromone profile reaches steady state, a cell that secretes pheromone globally would generate a local pheromone concentration at the surface of a neighboring cell of only 0.5 nM (Materials and Methods). However, if pheromone were secreted in a focused manner, the local concentration could exceed 5 nM, comparable to the receptor Kd (Jenness and Spatrick, 1986).
To better understand how a “local sensing” cell that detects pheromone at a zone surrounding the polarity site would respond to an adjacent partner, we simulated an arrangement with two spheres: a pheromone emitter and a pheromone receiver. The spheres were 250 nm apart (the minimal possible distance based on the combined thickness of two cell walls) to simulate cells that are touching (Figure 3A). α-Factor is secreted by exocytosis of vesicles, which fuse at a rate of ∼0.83/s (Dyer et al., 2013). Thus, with an overall α-factor release rate of 1400/s (Rogers et al., 2012), the average number of pheromone molecules in a vesicle would be ∼1680.
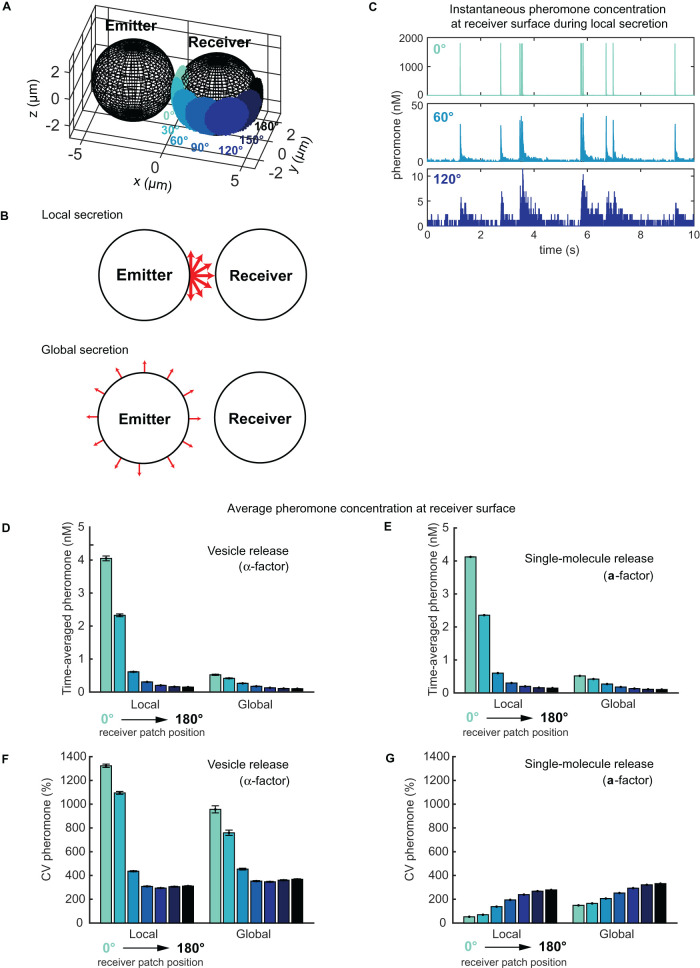
FIGURE 3:
Simulations of the pheromone receiver’s landscape for two touching cells. (A) Model setup for emitter and receiver cells shown at scale. Seven patch positions on the receiver (0° to 180°, changing colors) were used to measure local pheromone concentrations. (B) Local vs. global secretion. In local secretion, pheromone was released at the emitter pole abutting the receiver. In global secretion, pheromone was released uniformly at the emitter surface. (C) Instantaneous pheromone concentration at different positions (color) near receiver’s surface over time during local vesicle secretion. (D, E) Time-averaged pheromone concentration at different positions (color) on receiver’s surface for both vesicle and single-molecule release. (F, G) Coefficient of variation (CV) for D and E. All bars show mean ± SEM, n = 300 realizations.
We simulated pheromone release in one of two patterns: global secretion, where each vesicle releases its pheromone at a random position on the surface of the emitter, or local secretion, where each vesicle releases pheromone at the pole that abuts the receiver (Figure 3B). Following secretion, pheromone molecules were assumed to diffuse freely unless reflected from the surfaces of the two spheres. To simulate pheromone sensing in the vicinity of the polarity site, we designated a ∼1.3-μm-diameter patch at several locations (0° to 180°, changing colors, Figure 3A) on the receiver, and counted the number of molecules within 0.25 μm of the patch surface. Pheromone concentrations calculated in this manner fluctuated dramatically as vesicles were released (Figure 3C and Supplemental Figure S2).
Pheromone-receptor binding and unbinding are slow (koff = 0.01–0.001/s; Jenness et al., 1983; Raths et al., 1988; Yi et al., 2003; Bajaj et al., 2004), and, therefore, receptors are unable to respond rapidly to transient spikes in pheromone concentration. This suggests that receptors would time average the local concentration. Temporal averaging of simulated pheromone concentrations in different patches on the receiver cell indicated that the concentration sensed in the patch facing the emitter was approximately eightfold higher for simulations with polarized secretion compared with those with global secretion, consistent with the estimates discussed above (Figure 3D). Thus, if a threshold concentration must be detected to promote commitment, then cells that secrete pheromone in a polarized manner would be much more likely to cross that threshold. Furthermore, the pheromone concentrations sensed at different locations declined much more rapidly with distance from the emitter in the simulations with polarized secretion (Figure 3D). The steeper gradients created by polarized secretion may also be important for locating a mating partner. Thus, when secretion and sensing both occur at polarity sites, the concentration sensed by a cell would depend on the relative positions of the two cells’ polarity sites, as posited by the exploratory polarization hypothesis.
Unlike α-factor, a-factor is exported by the transporter Ste6 (Michaelis and Barrowman, 2012), so that a-factor release may occur one molecule at a time (Michaelis and Barrowman, 2012), rather than in vesicular packets. We repeated the simulations assuming the same overall production rate but releasing one molecule of pheromone at a time. While the variability in pheromone concentration was greatly reduced (Figure 3G, compare with F), the average pheromone concentrations sensed at different locations remained the same (Figure 3E, compare with D). In summary, local pheromone secretion would lead to approximately eightfold higher pheromone levels at the interface between partners.
Wild-type cells fail to commit to constitutively indecisive partners
Our findings indicate that pheromone secretion might be polarized during the indecisive phase, and that if it is, that would significantly alter local pheromone levels. But is polarized secretion important for communication to a partner cell? If pheromone emission is mainly global, then partner cells would commit to a partner regardless of that partner’s polarity site behavior. On the other hand, if pheromone emission is mainly local, the behavior of the polarity sites would be critical, and a cell would only commit to a partner that was reciprocating by stabilizing its own polarity site. To distinguish between these models, we explored the behavior of wild-type cells exposed to mutant partners that display constitutively indecisive polarity sites.
To quantify polarity site behavior, we developed an unbiased “spatial autocorrelation” scoring method to distinguish indecisive and committed polarity sites based on the Bem1 probe (Supplemental Figure S3A). We calculated the correlation between the spatial distribution of pixel intensities in a cell in consecutive time points. When polarity sites are mobile, the correlation is low, but if the cells commit, the correlation is high. We selected an autocorrelation threshold to designate commitment (Supplemental Figure S3B). In wild-type mating mixes where cells committed and fused with partners (Figure 4A and Supplemental Figure S4A), 19/20 a cells and 48/50 α cells crossed the threshold (Figure 4, C and E, and Supplemental Figure S3, C and D).
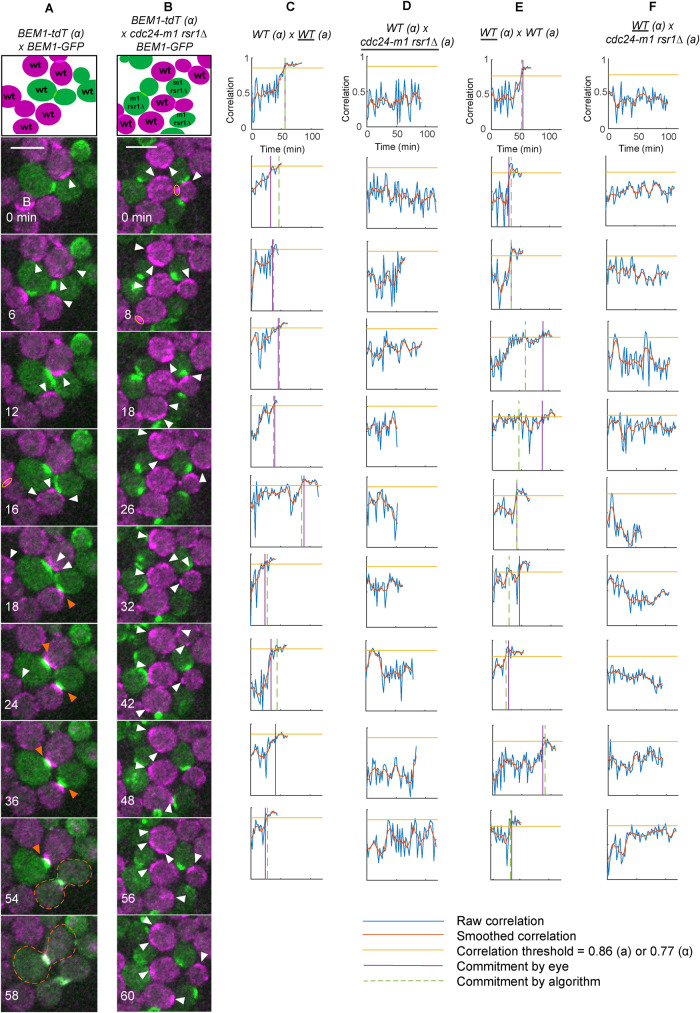
FIGURE 4:
Wild-type cells do not commit to mutants with constitutively mobile polarity sites. (A, B) Selected time points from movies of mating mixes imaged at 30°C. Cartoons indicate cells in the selected montages at the start of the displayed imaging interval. Yellow oval: mother–bud neck. White arrowhead: weak mobile Bem1-tdTomato clusters in wild-type partners. Orange arrowheads: stably oriented Bem1 clusters characteristic of committed cells. Dashed outline: fused zygote. (A) MATα wild-type cells (DLY12944; magenta) mixed with MATa wild-type cells (DLY9069; green). (B) MATα wild-type cells (DLY12944; magenta) mixed with MATa mutants that form constitutively mobile polarity clusters (cdc24-m1 rsr1Δ, DLY22797; green). (C, D) Spatial autocorrelation traces of representative wild-type (C) or cdc24-m1 rsr1Δ (D) cells mixed with wild-type α partners. Traces represent cells with the underlined genotype (a). (E, F) Spatial autocorrelation traces of representative wild-type α cells mixed with wild-type (E) or cdc24-m1 rsr1Δ (F) partners. Horizontal yellow line: threshold autocorrelation used to call commitment. Purple vertical line: commitment time as scored visually. Wild-type cells attempting to mate with constitutively indecisive mutants did not reach the threshold.
Polarity site stabilization (commitment) occurs by two parallel pathways, each of which recruits the Cdc42-directed guanine nucleotide exchange factor Cdc24 to the cortex (Dyer et al., 2013). One pathway depends on binding of the scaffold Far1 to Cdc24, which is impaired in the cdc24-m1 mutant (Valtz et al., 1995; Butty et al., 1998; Nern and Arkowitz, 1998, 1999). The other depends on the Ras-family GTPase Rsr1 (Bender and Pringle, 1989; Chant and Herskowitz, 1991). When treated with concentrated pheromone, mutants lacking both of these pathways (cdc24-m1 rsr1Δ) exhibit constitutively mobile polarity sites (Nern and Arkowitz, 2000; Dyer et al., 2013), and we found that they also exhibited constitutively mobile sites in a mating mix, with spatial autocorrelation remaining low (Figure 4, B and D, and Supplemental Figure S4B). Consistent with earlier work (Nern and Arkowitz, 1999), mating mixes between wild-type and cdc24-m1 rsr1Δ strains showed very poor mating efficiency (1/274 cells mated in mutant mix, n = 2 movies, but 305/503 cell mated in wild-type mix, n = 4 movies).
In control mating mixes with wild-type a cells, wild-type α cells committed to a partner after a variable indecisive phase (magenta cells in Figure 4A and Supplemental Movie 1). However, when mixed with cdc24-m1 rsr1Δ a cells, wild-type α cells failed to commit, instead exhibiting prolonged indecisive behavior (magenta cells in Figure 4B and Supplemental Movie 2). Similar results were obtained when wild-type a cells were mixed with cdc24-m1 rsr1Δ α cells (Supplemental Figure S4). Unlike in wild-type mating mixes, in mating mixes with cdc24-m1 rsr1Δ partners 0/20 wild-type cells (α) crossed the spatial autocorrelation threshold (Figure 4F and Supplemental Figure S3E). We conclude that wild-type cells cannot commit to a constitutively indecisive partner.
Movie S1.
Mating between cells harboring BEM1–GFP (DLY9069) and BEM1–tdTomato (DLY12944). Denoised maximum projections. Time in h:min. White arrowheads: selected polarity sites; orange arrowhead: commitment; dashed orange outline: zygote. Scale bar: 5 μm.
Movie S2.
Unsuccessful mating attempt between cells harboring cdc24–m1 rsr1Δ (DLY22797) and BEM1–tdTomato (DLY12944). Denoised maximum projections. Time in h:min. White arrowheads: initial polarity sites. Scale bar: 5 μm.
A potential caveat to our conclusion was that the failure of wild-type cells to commit to cdc24-m1 rsr1Δ mutants might be due to the mutants secreting less pheromone than wild-type cells. To measure pheromone secretion, we used the halo assay, in which cells from one mating type are deposited onto a spot over a lawn of cells of the opposite mating type. As the cells in the broader lawn proliferate, the pheromone released from the spotted cells causes cell-cycle arrest of nearby cells in the lawn, creating a “halo” (no-growth zone) whose diameter reflects the amount of pheromone released (see Materials and Methods; Manney, 1983). Halo assays indicated that cdc24-m1 rsr1Δ cells secreted comparable levels of pheromone to wild-type cells (both a-factor and α-factor; Supplemental Figure S5). These findings suggest that commitment by one partner requires reciprocal commitment by the other, consistent with the exploratory polarization hypothesis.
Behavior of wild-type cells mixed with prematurely committed partners
If wild-type cells cannot commit to partners that are constitutively indecisive, what about partners that commit prematurely? Addition of saturating levels of α-factor to a mating mix causes the a partner to polarize stably. As α-factor is everywhere, the a cell is “confused,” and polarizes in a random direction relative to the partner, resulting in a significant reduction of mating efficiency (Dorer et al., 1995, 1997). It is not known whether successful mating events in these conditions reflects a “unilateral” mating in which only one partner needs to orient properly, or a fortuitous coorientation between a “confused” a cell and an α partner.
We imaged wild-type cells in a mating mix with 10 μM α-factor. a cells polarized stably and grew in a single direction which usually did not point to an α partner. The α cells next to such “misoriented” a cells exhibited prolonged indecisive behavior and did not commit or mate (Figure 5A and Supplemental Movie 3). In the rarer instances in which an a cell polarized toward an α partner, some α cells polarized toward the a cell’s polarity site and mated (Figure 5B and Supplemental Movie 4). Thus, cells can mate with partners that stably orient in the correct direction, even if that orientation develops by chance and not through a search process. This accounts for all of the mating events we observed (n = 14). Interestingly, we also observed instances where pairs that appeared to be properly cooriented failed to mate (n = 36; Figure 5C and Supplemental Movie 5). The basis for this behavior remains unknown. Curiously, cells were able to mate even if the polarity site of the α partner was less stable than in typical pairings, as reflected in the fact that the spatial autocorrelation metric did not reach the commitment threshold (Figure 5D). Cells that did not mate (regardless of orientation) never reached the commitment threshold (Figure 5D). We did not observe any fusion events without preceding coorientation, suggesting that only if the two partners’ polarity sites are oriented toward each other do the cells commit and mate.
FIGURE 5:
Mating in “pheromone confusion” conditions. (A–C) Selected time points from movies of wild-type by wild-type mating mixes in the presence of 10 μM α-factor. MATα cells (DLY12944; magenta) were mixed with MATa cells (DLY9069; green; “confused” by excess pheromone). Confused cells polarize stably and form mating projections. White arrowhead: weak mobile Bem1 clusters in the MATα partners (magenta). (A) When MATa cell projections are oriented away from a potential partner, the partner’s polarity sites remain indecisive. (B) When MATa cell projections are oriented toward a potential partner, the cells can mate. (C) Mating can fail despite apparently correct orientation by the confused partner. (D) Representative spatial autocorrelation traces of MATα cells from instances of the categories illustrated in A–C.
Movie S3.
Unsuccessful mating attempt between cells harboring BEM1–GFP (DLY9069, MATa, “confused”) and BEM1–tdTomato (DLY12944, MATα, searching) plus 10 μM α–factor. Denoised maximum projections. Time in h:min. White arrowheads: initial polarity sites. Scale bar: 5 μm.
Movie S4.
Mating between cells harboring BEM1–GFP (DLY9069, MATa, “confused”) and BEM1–tdTomato (DLY12944, MATα, searching) plus 10 μM α–factor. Denoised maximum projections. Time in h:min. White arrowheads: selected polarity sites; orange arrowhead: commitment; dashed orange outline: zygote. Scale bar: 5 μm.
Movie S5.
Unsuccessful mating attempt between cells harboring BEM1–GFP (DLY9069, MATa, “confused”) and BEM1–tdTomato (DLY12944, MATα, searching) plus 10 μM α–factor. Denoised maximum projections. Time in h:min. White arrowheads: selected polarity sites. Scale bar: 5 μm.
Wild-type cells fail to commit to partners that lack polarity sites
Our findings thus far suggest that pheromone is emitted locally at polarity sites even during the indecisive phase, and that local emission at cooriented polarity sites is required in order to trigger commitment. If that is the case, then a cell that secreted pheromone globally would be unable to trigger commitment in its partner. To test that prediction, we wished to ask how wild-type cells respond to partners that cannot polarize Cdc42.
We first considered cells unable to activate Cdc42. At the restrictive temperature, cdc24-4ts mutants fail to polarize and have predominantly inactive Cdc42-GDP (Adams et al., 1990; Atkins et al., 2013). However, cdc24-4ts mutants also fail to activate the Cdc42 effector kinase Ste20 and cannot respond to pheromone (Simon et al., 1995; Zhao et al., 1995), so we introduced the constitutive ste20ΔCRIB in an attempt to restore pheromone signaling (Moskow et al., 2000). At restrictive temperature, wild-type cells failed to commit to unpolarized cdc24-4ts ste20ΔCRIB mutants (Supplemental Figure S6), consistent with the hypothesis that global pheromone emission is not sufficient to trigger commitment. However, interpretation of this result was complicated by the observation that cdc24-4ts ste20ΔCRIB mutants often failed to maintain G1 arrest (Supplemental Figure S6). We were able to enforce G1 arrest of the mutants (mating type a) by adding 10 μM α-factor. Pretreatment with α-factor at restrictive temperature caused the mutants to arrest as large unpolarized cells in G1 (see below). In mating mixes, wild-type α partners adjacent to these unpolarized mutants were efficiently arrested in G1 (91% of cells adjacent to a mutant remained unbudded for the median time of arrest in wild-type mixes, n = 142 cells, 3 movies). However, we did not observe mating between wild-type and mutant cells (0/83 cells mated to mutants, compared with 92/115 cells mated to wild-type partners at 35°C).
Wild-type cells committed to other wild-type cells (green cells in Figure 6A), but when mixed with unpolarized cdc24-4ts ste20ΔCRIB mutants, they failed to commit and exhibited prolonged indecisive behavior (green cells in Figure 6B and Supplemental Movie 6). We could not use the same spatial autocorrelation threshold for analysis of cells at 35°C (Figure 6B) because polarity sites were less bright and more mobile at 35°C than 30°C. Based on wild-type mating mixes at 35°C, we chose a different threshold to score commitment (Supplemental Figure S7, A and B). Using that threshold, 42/50 wild-type cells committed in matings with wild-type partners, while only 5/20 wild-type cells committed in mating mixes with cdc24-4ts ste20ΔCRIB partners (Figure 6D and Supplemental Figure S7C). These findings suggest that wild-type cells cannot commit to a partner lacking GTP-Cdc42.
FIGURE 6:
Wild-type cells do not commit to unpolarized partners. Selected time points from movies of mating mixes. B, bud; yellow oval, mother–bud neck. White arrowhead: weak, mobile clusters in wild-type partners. Orange arrowheads: stably oriented Bem1 clusters characteristic of committed cells. Dashed outline: fused zygote. 10 μM α-factor was added to B and C to sustain G1 arrest of mutant MATa cells. (A) MATα wild-type cells (DLY9070; green) mixed with MATa wild-type cells (DLY12943; magenta), imaged at 37°C. (B) The same MATα wild-type strain mixed with cdc24-4ts ste20ΔCRIB MATa cells (DLY23256; nuclear accumulation of Whi5-GFP, green, indicates G1 cells), imaged at 35°C. (C) MATα wild-type cells (DLY12944; magenta) mixed with MATa cells harboring membrane-targeted, constitutively active Cdc24 (MT-GFP-CDC2438A, DLY23351) that do not make polarity clusters. Scale bars: 5 µm. (D) Example spatial autocorrelation traces from wild-type cells mixed with the indicated partners. Horizontal yellow line: threshold autocorrelation used to call commitment. Purple vertical line: commitment time as scored visually. Wild-type cells attempting to mate with unpolarized mutants did not reach the threshold, even after 100 min.
Movie S6.
Unsuccessful mating attempt between cells harboring cdc24–4ts (DLY23256) and BEM1–GFP (DLY9070). Denoised maximum projections. Time in h:min. White arrowheads: initial polarity sites. Scale bar: 5 μm.
Because loss of Cdc42-GTP may have consequences beyond the absence of polarity sites, we also sought to eliminate polarity sites by activating Cdc42 all over the cortex. To that end, we overexpressed a membrane-targeted, constitutively active Cdc24 (MT-GFP-CDC2438A), a strategy previously shown to abrogate polarization (Kuo et al., 2014). Induction of membrane-targeted Cdc2438A blocks budding but allows cell-cycle progression until G2, where cells arrest due to the morphogenesis checkpoint (Lew, 2003). To ensure that the mutants (mating type a) were arrested in G1, we added 10 μM α-factor. In arrested mutant cells, both Bem1-tdTomato and MT-GFP-Cdc2438A were broadly distributed on the plasma membrane (Supplemental Figure S8). In mating mixes, wild-type α partners adjacent to these unpolarized mutants arrested efficiently in G1 (94% of cells adjacent to a mutant remained unbudded for the median duration of arrest in wild-type mixes, n = 116 cells, 3 movies). However, wild-type and mutant cells did not mate, even when imaged for 3 h (0/151 cells mated to mutants, compared with 100/162 cells mated to wild-type partners).
When mixed with MT-GFP-CDC2438A mutants, the wild-type α cells did not commit, instead displaying extended indecisive behavior (Figure 6C and Supplemental Movie 7). Whereas 48/50 cells mixed with wild-type partners developed spatial autocorrelation above threshold, 0/20 cells mixed with MT-CDC2438A partners did so, despite prolonged imaging (Figure 6D and Supplemental Figure S4D). We conclude that indecisive polarity sites are critical to communicate a cell’s location to its partner, and that without such communication, a wild-type cell does not commit.
Movie S7.
Unsuccessful mating attempt between cells harboring MT–GFP–CDC2438A BEM1–tdTomato (DLY23351) and BEM1–tdTomato (DLY12944). Denoised maximum projections. Time in h:min. White arrowheads: initial polarity sites. Scale bar: 5 μm.
DISCUSSION
Exploratory polarization underlies partner selection in yeast mating
Our findings indicate that the transient polarity sites formed during the indecisive period are critical for subsequent commitment to a mating partner. Proteins involved in pheromone sensing, secretion, and signaling were all enriched at these sites, suggesting that they are preferred sites for both pheromone secretion and sensing. Wild-type cells in mating mixes did not commit to partners that lacked polarity sites, partners with constitutively mobile polarity sites, or partners with stable but misoriented polarity sites. These results are fully consistent with the exploratory polarization hypothesis (Figure 1D), in which transient polarity sites mediate communication between mating partners.
Previous findings indicated that the appearance of a strong, stable polarity site, which we call “commitment,” results from detection of a high concentration of pheromone (Moore, 1983; Hegemann et al., 2015; McClure et al., 2015; Henderson et al., 2019). Moreover, MAPK activity increases concomitant with commitment to a partner (Conlon et al., 2016; Aymoz et al., 2018; Henderson et al., 2019), consistent with the idea that cells sense higher levels of pheromone when their polarity sites align. Our findings suggest that pheromone levels sufficient to trigger commitment are only achieved when a partner’s polarity site is directly apposed to that in the receiving cell. Simulations confirm that local pheromone secretion would expose a well-oriented polarity site to much higher pheromone levels than those attainable by a cell secreting pheromone uniformly around its surface. Thus, yeast cells may commit to a partner in response to the concentrated pheromone signal that accompanies coorientation of the two cells’ polarity sites.
An open question concerns the mechanism whereby the two partner cells’ polarity sites “find each other” to become cooriented. One could imagine that polarity sites form, move, and disappear stochastically until coorientation promotes stable commitment, as proposed for “speed dating” in S. pombe (Bendezu and Martin, 2013; Merlini et al., 2016). Alternatively, polarity sites may be guided toward each other by pheromone gradients. Our simulations indicate that when pheromone is secreted locally, the mating partner would experience a steep gradient in pheromone concentration, potentially guiding the movement or formation of polarity sites.
Reevaluating the pheromone landscape of mating cells
The observation that yeast cells are able to orient polarization toward artificial pheromone sources generated by micropipettes (Segall, 1993; Valtz et al., 1995; Nern and Arkowitz, 1998) or microfluidic devices (Paliwal et al., 2007; Hao et al., 2008; Moore et al., 2008, 2013; Jin et al., 2011; Brett et al., 2012; Lee et al., 2012; Dyer et al., 2013; Hegemann et al., 2015; Kelley et al., 2015; Vasen et al., 2020) has focused attention on the mechanism whereby cells decode a stable gradient of pheromone. Although yeast cells are clearly capable of polarizing growth toward an exogenous pheromone source, wild-type cells failed to polarize growth toward partners that were secreting pheromone uniformly around their surface. As such cells would be expected to set up a stable pheromone gradient similar to that from a micropipette, why did their partners not commit?
Experiments that analyze polarization of a cells in artificial α-factor gradients generally focus on cells that remain arrested in G1 for prolonged periods (4–10 h). Cells that are further from the pheromone source arrest only transiently and then resume budding, and these cells are omitted from the analysis of directional polarization in the gradient. However, we suggest that this transiently arrested population may be the most relevant to the behavior of mating cells. For the a cells in our wild-type by wild-type matings at 30°C, we found that 11% (n = 523 cells, 5 movies) of cells went on to bud during a 2 h observation window. Note that this analysis excludes cells that were not directly adjacent to (touching) potential G1-phase mating partners. Thus, cells that do not mate are unlikely to remain arrested in G1 for many hours under these circumstances. The simplest explanation for the failure of cells to commit to unpolarized partners is that yeast cells simply do not secrete enough pheromone to recreate the kinds of gradients produced by microfluidics devices.
Unlike experimental settings with unidirectional gradients, yeast cells in physiological mating scenarios must often discriminate between two or more similarly distant pheromone sources (Taxis et al., 2005; McClure et al., 2018). Under those circumstances, stable pheromone gradients would seem unlikely, and the findings presented in this and other recent studies (Bendezu and Martin, 2013; Dyer et al., 2013; Merlini et al., 2016; Henderson et al., 2019; Wang et al., 2019) suggest that mating cells operate in the context of a fluctuating pheromone landscape quite unlike the stable gradients studied thus far. Fluctuations occur on several timescales. First, vesicular release of α-factor would generate dramatic subsecond spikes in pheromone concentration, because each vesicle contains very concentrated (∼4 mM) α-factor. With an estimated α-factor diffusion constant of 150 μm2/s, each spike would dissipate to low nanomolar levels well before the next spike, generating rapid fluctuations. Second, the movement of the polarity sites during the indecisive phase means that the source of pheromone would relocate on a minute timescale, shifting the local gradients. Third, on a several-minute timescale, the mating or budding of nearby cells would remove them as pheromone sources in the local environment. Thus, physiological pheromone gradients are likely to be transitory, at least until the partners commit to each other. We suggest that the exploratory polarization strategy provides a framework for understanding how yeast cells are able to locate partners and mate successfully in such a dynamic pheromone landscape.
Advantages of exploratory polarization
The exploratory polarization strategy supported by our findings, like the related speed dating strategy proposed for S. pombe (Bendezu and Martin, 2013; Merlini et al., 2016; Martin, 2019), provides an elegant solution to the problem of choosing a partner from among two or more similarly distant candidates. Classical spatial sensing paradigms that integrate spatial information to extract a single “up-gradient” direction are poorly suited to this task, as the presence of two or more nearby chemoattractant sources may create a weak or even nonexistent net gradient. The task of picking just one of the potential partners is accomplished by the coincidence-detection feature of exploratory polarization: stabilization of the polarity site only occurs when the partners’ polarity sites happen to align (Figure 1D). By including this temporal aspect in the partner search process, the cells can avoid the potential paralysis that could ensue from access to two or more equally attractive partners. We note that this partner selection task occurs not only in mating, but also more broadly in multicellular contexts that involve the formation of focal cell–cell junctions like synapses.
A potential problem with exploratory polarization stems from the observation that during the indecisive phase, cells frequently developed two or more transient polarity sites. In principle, then, a cell could end up with two polarity sites, each oriented toward a different partner, leading to double mating. However, cells avoid this problem due to the competition between polarity sites that is built into the cell polarity circuit (Wu et al., 2015; Robertson et al., 2021). Because polarity factors are continually recruited into polarity clusters and then, after a few seconds, released back to the shared cytoplasm, states with more than one polarity site are transitory and only one polarity site can persist stably for long enough to promote fusion.
MATERIALS AND METHODS
Yeast strains and plasmids
Strains were constructed using standard molecular biology techniques. Yeast strains used in this study (Table 1) were generated in the YEF473 background (his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ63 ura3-52; Bi and Pringle, 1996), except DLY15660, which is in the 15D background (ade1, his2, leu2-3, 112 trp1-1, ura3Δns). The following alleles were previously described: BEM1-GFP:LEU2 (Kozubowski et al., 2008), BEM1-tdTomato:HIS3 (Howell et al., 2012), SPA2-mCherry:KANR (Howell et al., 2009), GFP-SEC4:URA3 (Chen et al., 2012), ura3:ste20(ΔCRIB):URA3 (Moran et al., 2019), STE2-sfGFP:URA3 and bar1Δ:URA3 (Henderson et al., 2019), rsr1Δ:HIS3 (Schenkman et al., 2002), STE5:GAL-STE5-CTM:GAL4BD-hER-VP16:LEU2 and SPA2-mCherry:HYGR (McClure et al., 2015), STE7(1-33)-NLS-NLS-mCherry:URA3 (Durandau et al., 2015), and GAL1p-PSR1(1-28)-GFP-CDC2438A:LEU2 (Woods et al., 2015).
Table 1:
Yeast and plasmid strains and genotypes.
| Yeast strain | Relevant genotype | Source |
|---|---|---|
| DLY8503 | MATα SPA2-mCherry:KANR | This study |
| DLY8993 | MATa bar1:URA3 | Henderson et al., 2019 |
| DLY9069 | MATa BEM1-GFP:LEU2 | Howell et al., 2009 |
| DLY9070 | MATα BEM1-GFP:LEU2 | Kozubowski et al., 2008 |
| DLY12943 | MATa BEM1-tdTomato:HIS3 | Henderson et al., 2019 |
| DLY12944 | MATα BEM1-tdTomato:HIS3 | This study |
| DLY13771 | MATa BEM1-tdTomato:HIS3 ura3:GFP-SEC4:URA3 | Henderson et al., 2019 |
| DLY15660 | MATα sst2:URA3 | This study |
| DLY20625 | MATα SPA2-mCherry:KANR STE5:GAL-STE5-CTM:GAL4BD-hER-VP16:LEU2 | This study |
| DLY20628 | MATa SPA2-mCherry:KANR WHI5-GFP:HIS5 STE5:GAL-STE5-CTM:GAL4BD-hER-VP16:LEU2 | This study |
| DLY20712 | MATa SPA2-mCherry:KANR STE2-sfGFP:URA3 | This study |
| DLY22243 | MATa BEM1-tdTomato:HIS3 STE2-sfGFP:URA3 | Henderson et al., 2019 |
| DLY22340 | MATα BEM1-tdTomato:HIS3 | Henderson et al., 2019 |
| DLY22355 | MATa BEM1-tdTomato STE6-sfGFP:KANR | This study |
| DLY22532 | MATa SPA2-mCherry:KANR WHI5-GFP:HIS5 STE5:GAL-STE5-CTM:GAL4BD-hER-VP16:LEU2 cdc24-m1:TRP1 rsr1Δ:HIS3 | This study |
| DLY22533 | MATα SPA2-mCherry:HYGR STE5:GAL-STE5-CTM:GAL4BD-hER-VP16:LEU2 cdc24-m1:TRP1 rsr1Δ:HIS3 | This study |
| DLY22797 | MATa BEM1-GFP:LEU2 cdc24-m1 (unmarked) rsr1Δ:HIS3 | This study |
| DLY23016 | MATa BEM1-GFP:LEU2 STE7(1-33)-NLS-NLS-mCherry:URA3 bar1::URA3 | This study |
| DLY23256 | MATa BEM1-tdTomato:HIS3 ura3:ste20(ΔCRIB):URA3 cdc24-4ts (unmarked) WHI5-GFP:HIS5 | This study |
| DLY23351 | MATa BEM1-tdTomato:HIS3 GAL1p-PSR1(1-28)-GFP-CDC2438A:LEU2 GAL-4BD-hER-VP16:URA3 WHI5-GFP:HIS5 | This study |
| DLY23354 | MATα BEM1-tdTomato:HIS3 GFP-STE4 (unmarked) | This study |
| DLY23612 | MATα BEM1-GFP:LEU2 cdc24-m1 (unmarked) rsr1Δ:HIS3 | This study |
| Plasmid | ||
|---|---|---|
| DLB52 | pFA6a-GFP(S65T)-HIS3MX6 | Bähler et al., 1998 |
| DLB3156 | pRS414-cdc24-m1 | Nern and Arkowitz, 1998 |
| DLB4171 | pRS41N-GFP-STE4 | This study |
| DLB4254 | pRSII306-Ste4prom-GFP-Ste4(1-194) | This study |
| DLB4292 | pFA6a-link-yoSuperfolderGFP-Kan | Lee et al., 2013 |
| DLB4435 | pRS306-cdc24-m1:URA3 | This study |
| DLB4287 | pRS304-cdc24-m1-C:TRP1 | This study |
The sst2Δ:URA3 disruption was generated by the one-step PCR-based method (Baudin et al., 1993) using pRS306 as template.
WHI5-GFP:S.p.HIS5 (Doncic et al., 2011) and STE6-sfGFP:KANR were constructed using methods described previously (Longtine et al., 1998) with DLB52 (pFA6a-GFP(S65T)-S.p.HIS5MX6; Addgene plasmid #41598) and DLB4292 (pFA6a-link-yoSuperfolderGFP-Kan; Addgene plasmid #44901) as templates.
GFP-STE4 was constructed in the YEF background by a pop-in, pop-out strategy. First, GFP-STE4 was PCR-amplified from a strain derived from RDY126 (Suchkov et al., 2010). This fragment was inserted into pRS41N (Taxis and Knop, 2006) using ApaI and NotI, producing DLB4171 (pRS41N-GFP-STE4). DLB4171 was digested with HindIII and ApaI to release a fragment containing the STE4 promoter, GFP, and base pairs 1–194 of the ORF. This fragment was inserted into DLB212 (pRSII306: a URA3-marked integrating plasmid) to produce DLB4254 (pRSII306-Ste4prom-GFP-Ste4(1–194)). DLB4254 was partially digested with PstI to target integration to the STE4 locus of a diploid from the YEF background. Haploid segregants containing STE4(1–194), URA3 marker, and GFP-STE4 at the STE4 locus were plated on medium containing 5-fluoroorotic acid to select for colonies in which recombination occurred between the promoter of the STE4 fragment and the promoter of GFP-STE4, removing the URA3 marker and leaving GFP-STE4 as a precise replacement of the endogenous STE4.
To introduce the cdc24-4 allele into the YEF473 strain background, we first deleted one copy of CDC24 in a diploid strain using the HIS3 marker. A centromeric URA3-marked plasmid carrying wild-type CDC24 was transformed into the strain, and following sporulation and tetrad dissection a haploid cdc24::HIS3 strain carrying the plasmid was selected. cdc24-4 was amplified by PCR from a strain derived from JPT19-H01 (Sloat et al., 1981), and used to replace the cdc24::HIS3 allele by homologous recombination, followed by selection on medium containing 5-fluoroorotic acid to obtain colonies without the plasmid.
The cdc24-m1 allele was amplified by PCR from pRS414-cdc24-m1 (Nern and Arkowitz, 1998) and cloned into pRS306 to produce DLB4435 (pRS306-cdc24-m1). DLB4435 was digested with BspEI to target integration at the CDC24 promoter, yielding a locus where the URA3 gene is inserted between the cdc24-m1 allele and wild-type CDC24. Haploid MATa segregants were grown on medium containing 5-fluoroorotic acid to select for recombination between cdc24-m1 and CDC24. Recombinants containing cdc24-m1 were identified by phenotyping (morphology when treated with pheromone) and confirmed by sequencing. To generate cdc24-m1:TRP1, DLB4287 containing a C-terminal fragment of cdc24-m1 ORF and 501 base pairs of 3′-UTR in the pRS304 backbone (Sikorski and Hieter, 1989) was digested at the unique XcmI site in cdc24-m1 to target integration at CDC24. This generates a tandem duplication with full-length cdc24-m1 followed by TRP1 and a promoterless truncated CDC24 fragment.
Live-cell microscopy
Cells were grown in complete synthetic medium (CSM; MP Biomedicals, Solon, OH) with 2% dextrose (Macron, Center Valley, PA) overnight at 30°C to midlog phase (106–107 cells/ml). Cultures of opposite mating type strains were mixed to obtain a 1:1 cell ratio, centrifuged to concentrate the cells, and mounted on CSM–dextrose slabs solidified with 2% agarose (Hoefer, Holliston, MA) and sealed with petroleum jelly. For pheromone “confusion” experiments, cells were imaged on a slab containing 10 μM α-factor (Genway Biotech, San Diego, CA). For uniform pheromone experiments, cells were imaged on a slab containing 5 nM α-factor. GAL1p-MT-CDC2438A expression was induced in dextrose medium by adding β-estradiol (Sigma-Aldrich, St. Louis, MO) to the medium to a final concentration of 20 nM, incubating for 2 h, and imaging on a slab containing 20 nM β-estradiol (the strains contain an artificial transcription factor, GAL-4BD-hER-VP16, that induces the GAL1 promoter in response to β-estradiol). For cdc24-4ts strains, cells were grown overnight at 24°C and shifted to 37°C for 2 h before imaging. For cells that were arrested in G1 (MT-CDC2438A or cdc24-4ts), α-factor was added to a final concentration of 10 μM, and cells were incubated for 2 h before imaging on a slab containing 10 μM α-factor. Imaging was performed in a temperature-controlled chamber at 30°C or the restrictive temperature as indicated.
Images were acquired with an Andor Revolution XD spinning-disk confocal microscope (Andor Technology, Concord, MA) with a CSU-X1 5000-rpm confocal scanner unit (Yokogawa, Tokyo, Japan) and a UPLSAPO 100× /1.4 oil-immersion objective (Olympus, Tokyo, Japan), controlled by MetaMorph software (Molecular Devices, San Jose, CA). Images were captured by an iXon3 897 EM-CCD camera with 1.2× auxiliary magnification (Andor Technology).
Images were acquired in z-stacks (15 0.47-μm steps) at 2-min intervals. Laser power varied by experiment but was set to levels that produced bright signals with minimal bleaching during the movie: 8–15% (488 nm) and 10–15% (561 nm) of maximal output. EM gain was 200, and exposure time was 250 ms. Images were denoised with the ImageJ Hybrid 3D Median Filter plug-in (2007), created by Christopher Philip Mauer and Vytas Bindokas. Images are maximum projections except for MT-CDC2438A medial plane image, as indicated. Scaling of images was always matched for experimental and relevant control conditions.
Scoring colocalization of Ste2, Ste4, Ste6, and Sec4 with Bem1
Scoring was conducted in two steps: 1) Selection of scorable time points. From mating movies, we identified all cells in G1 that were adjacent to at least one potential G1-phase partner. Time points with 1 or 2 predominant Bem1 clusters or arcs/crescents were selected for scoring. Arcs or crescents of uneven intensity were considered a single continuous region. Time points where the Bem1 signal was dim, diffusely distributed, covered more than 50% of the cortex, appeared in more than two clusters, or could otherwise not be clearly localized to one or two distinct regions were excluded from scoring. Bem1 signal at the neck (cytokinesis or cytokinesis remnant) was ignored, but neck-adjacent signal could be scored. Occasional tight puncta (less than five pixels) of Bem1 were also ignored. 2) Scoring colocalization. For each scorable time point, Bem1 and the protein of interest were considered colocalized if the signal of interest was more intense at/near the Bem1 signal than in the broader neighborhood. Because some other probes localized to tighter foci than Bem1, complete overlap was not required to score an image as colocalized. Signals at other, noncolocalized regions were also allowed. For time points with two comparable Bem1 clusters, overlap with either cluster was scored as colocalization. For time points with two unequal clusters, overlap with the more prominent cluster was required to score colocalization. Because Ste2-sfGFP often accumulated on the plasma membrane as a crescent, a region of Ste2 was judged to be colocalized only if the more intense part of the crescent overlapped with the Bem1 cluster. To minimize inherent subjectivity, cases judged ambiguous by the rater were excluded.
Mating efficiency
Mating efficiencies were calculated from mating movies. We tracked all cells in G1 that were adjacent to at least one potential G1-phase partner (i.e., “available” to mate) during a 2-h movie and scored them as “mated” or “not mated.” Cells whose potential partners mated with another cell and cells that were available to mate only during the last 20 min were excluded from the sample.
Budding index
Budding indices were calculated from mating movies. Using the appearance of Bem1-GFP or Bem1-tdTomato at the neck as a marker of G1, we first calculated the duration of G1 for all wild-type α cells that mated (mutants were a cells) and determined the median of that value (52 min). We then tracked all cells in G1 that were adjacent to at least one potential G1-phase partner (i.e., “available” to mate) for 52 min, noting whether they budded, mated, or remained arrested.
Halo assays to measure pheromone secretion
Halo assays to measure pheromone secretion. a-Factor pheromone promotes G1 arrest of MATα cells, while α-factor pheromone promotes G1 arrest of MATa cells. Halo assays detect the pheromone produced by a spot of cells placed on a lawn of supersensitive cells of the opposite mating type. Lawns (3.5 × 106 cells) were spread on YEPD plates (1% yeast extract, 2% peptone, 2% dextrose). Lawn strains were DLY8993 (MATa bar1Δ) to detect α-factor and DLY15660 (MATα sst2Δ) to detect a-factor. After spreading the lawn on the plate and allowing it to dry, spots containing 107 cells of the strains to be tested were pipetted onto the lawn. Halos (zones of G1 arrest) were photographed after 48 h at 30°C. Because cells make more pheromone when they are in G1 phase (Henderson et al., 2019), we performed similar assays using cells that were arrested in G1 by expression of Ste5-CTM (Pryciak and Huntress, 1998). Expression was induced by the addition of 300 nM β-estradiol to the plate. Halo diameter was quantified by fitting a circle to each halo.
Spatial autocorrelation analysis
The image processing toolbox in MATLAB 2019a was used to develop a custom tool to track individual cells during the mating period and determine commitment to a partner in an unbiased way. From mating movies, we identified wild-type α cells (BEM1-GFP or BEM1-tdTomato) that were available to mate with a-cell partners. We circumscribed the wild-type cells at several time points throughout the movie, beginning at G1 and ending either at the time point preceding fusion (for cells that mated) or the end of the movie (for cells that did not mate). Using linear interpolation, the outlines were deformed over time to accommodate changes in cell morphology and position. In this manner, we obtained cell outlines between the marked time points, enabling continuous tracking of each cell during the mating period.
The spatial array of intracellular Bem1 signal was extracted at each time point, and the correlation between arrays at adjacent time points was calculated (“spatial autocorrelation”), using the following formula:
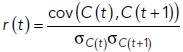 |
where r is Pearson’s coefficient, cove is covariance, C is an array containing indexed fluorescence data, t is time point, and σc is the SD of the array C. The two arrays C(t) and C(t + 1) were obtained by using a union of the outlines at time points t and t + 1 to ensure that spatial overlap was continuous.
During the indecisive period, r is relatively low, but at commitment, r is relatively high. To determine a threshold of r that indicates commitment, we performed a sweep through different threshold values for a set of cells for which an experienced rater had already judged the time of commitment. A threshold value was selected to minimize discrepancies (either early or late judgments) between the automated and the human rater. We note that different thresholds were selected for different probes (GFP or tdTomato) and different temperatures to account for imaging variations. The code used for this analysis can be found at https://github.com/DebrajGhose/Exploratory-polarization-yeast.
Calculation of pheromone concentrations expected for global and local secreting cells
To gain insight into the types of gradients expected from global and local secreting cells, we considered the gradient generated by a spherical emitter centered at the origin (Rappaport and Barkai, 2012). For this case, the pheromone gradient can be found by solving Laplace’s equation using spherical coordinates. The r-coordinate satisfies the equation
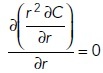 |
The boundary conditions are constant flux density J (number of molecules released per unit time per unit area) at the surface of the sphere (r = R)
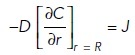 |
and the concentration vanishes as  With these boundary conditions, the concentration takes the following form:
With these boundary conditions, the concentration takes the following form:
In terms of the total secretion rate S, the above expression becomes
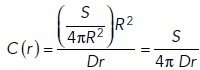 |
If we assume a secretion rate of 1400 molecules/s and diffusion coefficient for pheromone molecules of 150 μm2/s, then the concentration at the surface of cell of radius 2.5 μm is C(2.5) = 0.3 molecules/μm3 = 0.5 nM. If we assume the localized emitter has a radius of 0.25 μm, then the concentration at the surface of the emitter is 10 times higher C(0.25) = 5 nM. These results are consistent with the emitter alone results shown in Supplemental Figure S2B.
Simulations of pheromone landscape for two touching cells
Particle-based simulations of pheromone emission and diffusion were conducted using the Smoldyn software (v2.58) on Linux systems (2.50 GHz and 2.30 GHz Intel processors; Longleaf cluster at UNC Chapel Hill, NC; Andrews and Bray, 2004; Andrews, 2017). The code is available at https://github.com/mikepab/exploratory-polarization-yeast. Pheromone molecules were modeled as Brownian point particles with diffusion coefficient D = 150 µm2/s, and were removed at a spherical absorbing boundary 50 µm from the origin. A mating pair was modeled as two spheres, a receiver and emitter, centered at (± (2.5 + 0.25/2) µm, 0 µm, 0 µm) with radius 2.5 µm. The system was first equilibrated for 5 s, after which coordinates were recorded every 0.1 ms timestep for 10 s. For each condition, n = 300 realizations were conducted.
Vesicle emission events were simulated by repeated use of the Smoldyn command cmd @ t pointsource pheromone n x y z. First, t specifies the time of a single emission event; intervals between each emission were exponentially distributed:
pheromone is a molecular species defined in the Smoldyn script, n is the number of molecules released per vesicle (n = 1663), and x y z are spatial coordinates of the vesicle event. In the local case, (x = −0.25/2+0.001 µm, y = z = 0 µm). In the global case, the spatial coordinates were obtained by uniform random sampling on a sphere centered at (−(2.5+0.25/2) µm, 0 µm, 0 µm) with radius 2.5001 µm.
Single-molecule emission events were handled using the Smoldyn reaction surface = and reaction_production commands, with a release rate per timestep of 1/7.1429 (yielding 1400 molecules per second at 0.1 ms timesteps). In the local case, the releasing surface was a sphere centered at x = −0.25/2+0.001 µm, y = z = 0 µm with a radius of 0.0005 µm. In the global case, the releasing surface was a sphere centered at (−(2.5+0.25/2), 0, 0) µm with radius 2.5001 µm.
To validate the simulation setup, we set up simulations comparable to the analytic solution described above. The receiver was removed and pheromone profiles were measured as a function of distance to the emitter. The simulations were in good agreement with the analytic solution (Supplemental Figure S2B).
Analysis of particle-based pheromone simulations
Receiver-centered molecular coordinates were filtered to only include pheromone within 0.25 µm of the receiver surface. Then, a 3D angle between each molecule 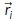 and a reference vector
and a reference vector 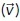 was calculated:
was calculated:
The reference vector defines the patch under consideration (Figure 3A). For 0°, the closest patch to the emitter 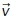 = [–1,0,0]. For the other patches, we rotate [–1,0,0] by the desired angle θrot.
= [–1,0,0]. For the other patches, we rotate [–1,0,0] by the desired angle θrot.
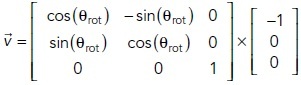 |
To count molecules in each patch, we summed the number of points within 0 ≤ θi ≤ 30°. The volume of each patch is 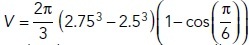 , which was used to convert molecules to nanomolar. Finally, a time-averaged pheromone concentration and coefficient of variation (CV) were calculated for each patch in each simulation, allowing us to compute a mean and standard error across simulations (Figure 3, D–G).
, which was used to convert molecules to nanomolar. Finally, a time-averaged pheromone concentration and coefficient of variation (CV) were calculated for each patch in each simulation, allowing us to compute a mean and standard error across simulations (Figure 3, D–G).
Supplementary Material
Acknowledgments
We thank Stefano Di Talia, Masayuki Onishi, and Amy Gladfelter, as well as members of the Lew lab, for stimulating conversations and comments on the manuscript, and Alec D’Alessandro for the data on cells in uniform pheromone. M.R.C.-C. was a Howard Hughes Medical Institute Gilliam Fellow and received a Graduate Diversity Enrichment Program Award from the Burroughs Wellcome Fund. This work was funded by National Institutes of Health/National Institute of General Medical Sciences Grants no. R35GM-127145 to T.C.E., and no. R01GM-103870 and no. R35GM-122488 to D.J.L.
Abbreviations used:
- GFP
green fluorescent protein
- GTPase
guanosine triphosphate hydrolase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-02-0068) on March 10, 2021.
REFERENCES
- Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR (1990). CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol 111, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SS (2017). Smoldyn: particle-based simulation with rule-based modeling, improved molecular interaction and a library interface. Bioinformatics 33, 710–717. [DOI] [PubMed] [Google Scholar]
- Andrews SS, Bray D (2004). Stochastic simulation of chemical reactions with spatial resolution and single molecule detail. Phys Biol 1, 137–151. [DOI] [PubMed] [Google Scholar]
- Arkowitz RA (1999). Responding to attraction: chemotaxis and chemotropism in Dictyostelium and yeast. Trends Cell Biol 9, 20–27. [DOI] [PubMed] [Google Scholar]
- Atkins BD, Yoshida S, Saito K, Wu CF, Lew DJ, Pellman D (2013). Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J Cell Biol 202, 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymoz D, Solé C, Pierre JJ, Schmitt M, de Nadal E, Posas F, Pelet S (2018). Timing of gene expression in a cell-fate decision system. Mol Syst Biol 14, e8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR, Drubin DG (1998). A role for the yeast actin cytoskeleton in pheromone receptor clustering and signalling. Curr Biol 8, 927–930. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast (Chichester, England) 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Bajaj A, Celic´ A, Ding FX, Naider F, Becker JM, Dumont ME (2004). A fluorescent α-factor analogue exhibits multiple steps on binding to its G protein coupled receptor in yeast. Biochemistry 43, 13564–13578. [DOI] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C (1993). A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellon A, Mann F (2018). Keeping up with advances in axon guidance. Curr Opin Neurobiol 53, 183–191. [DOI] [PubMed] [Google Scholar]
- Bender A, Pringle JR (1989). Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc Natl Acad Sci USA 86, 9976–9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu FO, Martin SG (2013). Cdc42 explores the cell periphery for mate selection in fission yeast. Curr Biol 23, 42–47. [DOI] [PubMed] [Google Scholar]
- Bi E, Pringle JR (1996). ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol 16, 5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett ME, DeFlorio R, Stone DE, Eddington DT (2012). A microfluidic device that forms and redirects pheromone gradients to study chemotropism in yeast. Lab Chip 12, 3127–3134. [DOI] [PubMed] [Google Scholar]
- Butty AC, Pryciak PM, Huang LS, Herskowitz I, Peter M (1998). The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science 282, 1511–1516. [DOI] [PubMed] [Google Scholar]
- Chant J, Herskowitz I (1991). Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65, 1203–1212. [DOI] [PubMed] [Google Scholar]
- Chen H, Kuo CC, Kang H, Howell AS, Zyla TR, Jin M, Lew DJ (2012). Cdc42p regulation of the yeast formin Bni1p mediated by the effector Gic2p. Mol Biol Cell 23, 3814–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon P, Gelin-Licht R, Ganesan A, Zhang J, Levchenko A (2016). Single-cell dynamics and variability of MAPK activity in a yeast differentiation pathway. Proc Natl Acad Sci USA 113, E5896–E5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Thorner JW (2001). Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem 70, 703–754. [DOI] [PubMed] [Google Scholar]
- Doncic A, Falleur-Fettig M, Skotheim JM (2011). Distinct interactions select and maintain a specific cell fate. Mol Cell 43, 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer R, Boone C, Kimbrough T, Kim J, Hartwell LH (1997). Genetic analysis of default mating behavior in Saccharomyces cerevisiae. Genetics 146, 39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer R, Pryciak PM, Hartwell LH (1995). Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J Cell Biol 131, 845–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durandau E, Aymoz D, Pelet S (2015). Dynamic single cell measurements of kinase activity by synthetic kinase activity relocation sensors. BMC Biol 13, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JM, Savage NS, Jin M, Zyla TR, Elston TC, Lew DJ (2013). Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr Biol 23, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Nayak S, Behar M, Shanks RH, Nagiec MJ, Errede B, Hasty J, Elston TC, Dohlman HG (2008). Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell 30, 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann B, Peter M (2017). Local sampling paints a global picture: local concentration measurements sense direction in complex chemical gradients. Bioessays 39, 201600134. [DOI] [PubMed] [Google Scholar]
- Hegemann B, Unger M, Lee SS, Stoffel-Studer I, van den Heuvel J, Pelet S, Koeppl H, Peter M (2015). A cellular system for spatial signal decoding in chemical gradients. Dev Cell 35, 458–470. [DOI] [PubMed] [Google Scholar]
- Henderson NT, Pablo M, Ghose D, Clark-Cotton MR, Zyla TR, Nolen J, Elston TC, Lew DJ (2019). Ratiometric GPCR signaling enables directional sensing in yeast. PLoS Biol 17, e3000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T, Takeuchi H (2015). The mechanism and key molecules involved in pollen tube guidance. Annu Rev Plant Biol 66, 393–413. [DOI] [PubMed] [Google Scholar]
- Howell AS, Jin M, Wu C-F, Zyla TR, Elston TC, Lew DJ (2012). Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell 149, 322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, Lew DJ (2009). Singularity in polarization: rewiring yeast cells to make two buds. Cell 139, 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismael A, Stone DE (2017). Yeast chemotropism: a paradigm shift in chemical gradient sensing. Cell Logist 7, e1314237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL, Hartwell LH (1990a). Courtship in S. cerevisiae: both cell types choose mating partners by responding to the strongest pheromone signal. Cell 63, 1039–1051. [DOI] [PubMed] [Google Scholar]
- Jackson CL, Hartwell LH (1990b). Courtship in Saccharomyces cerevisiae: an early cell-cell interaction during mating. Mol Cell Biol 10, 2202–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenness DD, Burkholder AC, Hartwell LH (1983). Binding of α-factor pheromone to yeast a cells: chemical and genetic evidence for an α-factor receptor. Cell 35, 521–529. [DOI] [PubMed] [Google Scholar]
- Jenness DD, Spatrick P (1986). Down regulation of the α-factor pheromone receptor in S. cerevisiae. Cell 46, 345–353. [DOI] [PubMed] [Google Scholar]
- Jin M, Errede B, Behar M, Mather W, Nayak S, Hasty J, Dohlman HG, Elston TC (2011). Yeast dynamically modify their environment to achieve better mating efficiency. Sci Signal 4, ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Jin M, Lew DJ (2011). Symmetry breaking and the establishment of cell polarity in budding yeast. Curr Opin Genet Dev 21, 740–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JB, Dixit G, Sheetz JB, Venkatapurapu SP, Elston TC, Dohlman HG (2015). RGS proteins and septins cooperate to promote chemotropism by regulating polar cap mobility. Curr Biol 25, 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ (2008). Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr Biol 18, 1719–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C-C, Savage NS, Chen H, Wu C-F, Zyla TR, Lew DJ (2014). Inhibitory GEF phosphorylation provides negative feedback in the yeast polarity circuit. Curr Biol 24, 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Horvath P, Pelet S, Hegemann B, Lee LP, Peter M (2012). Quantitative and dynamic assay of single cell chemotaxis. Integr Biol (Camb) 4, 381–390. [DOI] [PubMed] [Google Scholar]
- Lee S, Lim WA, Thorn KS (2013). Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae. PLoS One 8, e67902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ (2003). The morphogenesis checkpoint: how yeast cells watch their figures. Curr Opin Cell Biol 15, 648–653. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast (Chichester, England) 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Manney TR (1983). Expression of the BAR1 gene in Saccharomyces cerevisiae: induction by the alpha mating pheromone of an activity associated with a secreted protein. J Bacteriol 155, 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG (2019). Molecular mechanisms of chemotropism and cell fusion in unicellular fungi. J Cell Sci 132, jcs230706. [DOI] [PubMed] [Google Scholar]
- McClure AW, Jacobs KC, Zyla TR, Lew DJ (2018). Mating in wild yeast: delayed interest in sex after spore germination. Mol Biol Cell 29, 3119–31278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure AW, Minakova M, Dyer JM, Zyla TR, Elston TC, Lew DJ (2015). Role of polarized G protein signaling in tracking pheromone gradients. Dev Cell 35, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L, Dudin O, Martin SG (2013). Mate and fuse: how yeast cells do it. Open Biol 3, 130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L, Khalili B, Bendezu FO, Hurwitz D, Vincenzetti V, Vavylonis D, Martin SG (2016). Local pheromone release from dynamic polarity sites underlies cell-cell pairing during yeast mating. Curr Biol 26, 1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis S, Barrowman J (2012). Biogenesis of the Saccharomyces cerevisiae pheromone a-factor, from yeast mating to human disease. Microbiol Mol Biol Rev 76, 626–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SA (1983). Comparison of dose-response curves for alpha factor-induced cell division arrest, agglutination, and projection formation of yeast cells. Implication for the mechanism of alpha factor action. J Biol Chem 258, 13849–13856. [PubMed] [Google Scholar]
- Moore TI, Chou C-S, Nie Q, Jeon NL, Yi T-M (2008). Robust spatial sensing of mating pheromone gradients by yeast cells. PLoS One 3, e3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore TI, Tanaka H, Kim HJ, Jeon NL, Yi T-M (2013). Yeast G-proteins mediate directional sensing and polarization behaviors in response to changes in pheromone gradient direction. Mol Biol Cell 24, 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran KD, Kang H, Araujo AV, Zyla TR, Saito K, Tsygankov D, Lew DJ (2019). Cell-cycle control of cell polarity in yeast. J Cell Biol 218, 171–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskow JJ, Gladfelter AS, Lamson RE, Pryciak PM, Lew DJ (2000). Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol Cell Biol 20, 7559–7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Arkowitz RA (1998). A GTP-exchange factor required for cell orientation. Nature 391, 195–198. [DOI] [PubMed] [Google Scholar]
- Nern A, Arkowitz RA (1999). A Cdc24p-Far1p-Gβγ protein complex required for yeast orientation during mating. J Cell Biol 144, 1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Arkowitz RA (2000). G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol Cell 5, 853–864. [DOI] [PubMed] [Google Scholar]
- Nichols JM, Veltman D, Kay RR (2015). Chemotaxis of a model organism: progress with Dictyostelium. Curr Opin Cell Biol 36, 7–12. [DOI] [PubMed] [Google Scholar]
- Paliwal S, Iglesias PA, Campbell K, Hilioti Z, Groisman A, Levchenko A (2007). MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature 446, 46–51. [DOI] [PubMed] [Google Scholar]
- Park HO, Bi E (2007). Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev 71, 48–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A (2004). Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol 20, 559–591. [DOI] [PubMed] [Google Scholar]
- Pryciak PM, Huntress FA (1998). Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev 12, 2684–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport N, Barkai N (2012). Disentangling signaling gradients generated by equivalent sources. J Biol Phys 38, 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raths SK, Naider F, Becker JM (1988). Peptide analogues compete with the binding of alpha-factor to its receptor in Saccharomyces cerevisiae. J Biol Chem 263, 17333–17341. [PubMed] [Google Scholar]
- Robertson CG, Clark-Cotton MR, Lew DJ (2021). Mechanisms that ensure monogamous mating in Saccharomyces cerevisiae. Mol Biol Cell 32, 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers DW, McConnell E, Greig D (2012). Molecular quantification of Saccharomyces cerevisiae α-pheromone secretion. FEMS Yeast Res 12, 668–674. [DOI] [PubMed] [Google Scholar]
- Sarris M, Sixt M (2015). Navigating in tissue mazes: chemoattractant interpretation in complex environments. Curr Opin Cell Biol 36, 93–102. [DOI] [PubMed] [Google Scholar]
- Schenkman LR, Caruso C, Pagé N, Pringle JR (2002). The role of cell cycle-regulated expression in the localization of spatial landmark proteins in yeast. J Cell Biol 156, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall JE (1993). Polarization of yeast cells in spatial gradients of alpha mating factor. Proc Natl Acad Sci USA 90, 8332–8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Geneticsl 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MN, De Virgilio C, Souza B, Pringle JR, Abo A, Reed SI (1995). Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376, 702–705. [DOI] [PubMed] [Google Scholar]
- Sloat BF, Adams A, Pringle JR (1981). Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J Cell Biol 89, 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchkov DV, DeFlorio R, Draper E, Ismael A, Sukumar M, Arkowitz R, Stone DE (2010). Polarization of the yeast pheromone receptor requires its internalization but not actin-dependent secretion. Mol Biol Cell 21, 1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Keller P, Kavagiou Z, Jensen LJ, Colombelli J, Bork P, Stelzer EHK, Knop M (2005). Spore number control and breeding in Saccharomyces cerevisiae: a key role for a self-organizing system. J Cell Biol 171, 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Knop M (2006). System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. Biotechniques 40, 73–78. [DOI] [PubMed] [Google Scholar]
- Valtz N, Peter M, Herskowitz I (1995). FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J Cell Biol 131, 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasen G, Dunayevich P, Colman-Lerner A (2020). Mitotic and pheromone-specific intrinsic polarization cues interfere with gradient sensing in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 117, 6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tian W, Banh BT, Statler B-M, Liang J, Stone DE (2019). Mating yeast cells use an intrinsic polarity site to assemble a pheromone-gradient tracking machine. J Cell Biol 218, 3730–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods B, Kuo C-C, Wu C-F, Zyla TR, Lew DJ (2015). Polarity establishment requires localized activation of Cdc42. J Cell Biol 211, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CF, Chiou JG, Minakova M, Woods B, Tsygankov D, Zyla TR, Savage NS, Elston TC, Lew DJ (2015). Role of competition between polarity sites in establishing a unique front. Elife 4, e11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi TM, Kitano H, Simon MI (2003). A quantitative characterization of the yeast heterotrimeric G protein cycle. Proc Natl Acad Sci USA 100, 10764–10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZS, Leung T, Manser E, Lim L (1995). Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator CDC24. Mol Cell Biol 15, 5246–5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.