Figure 2.
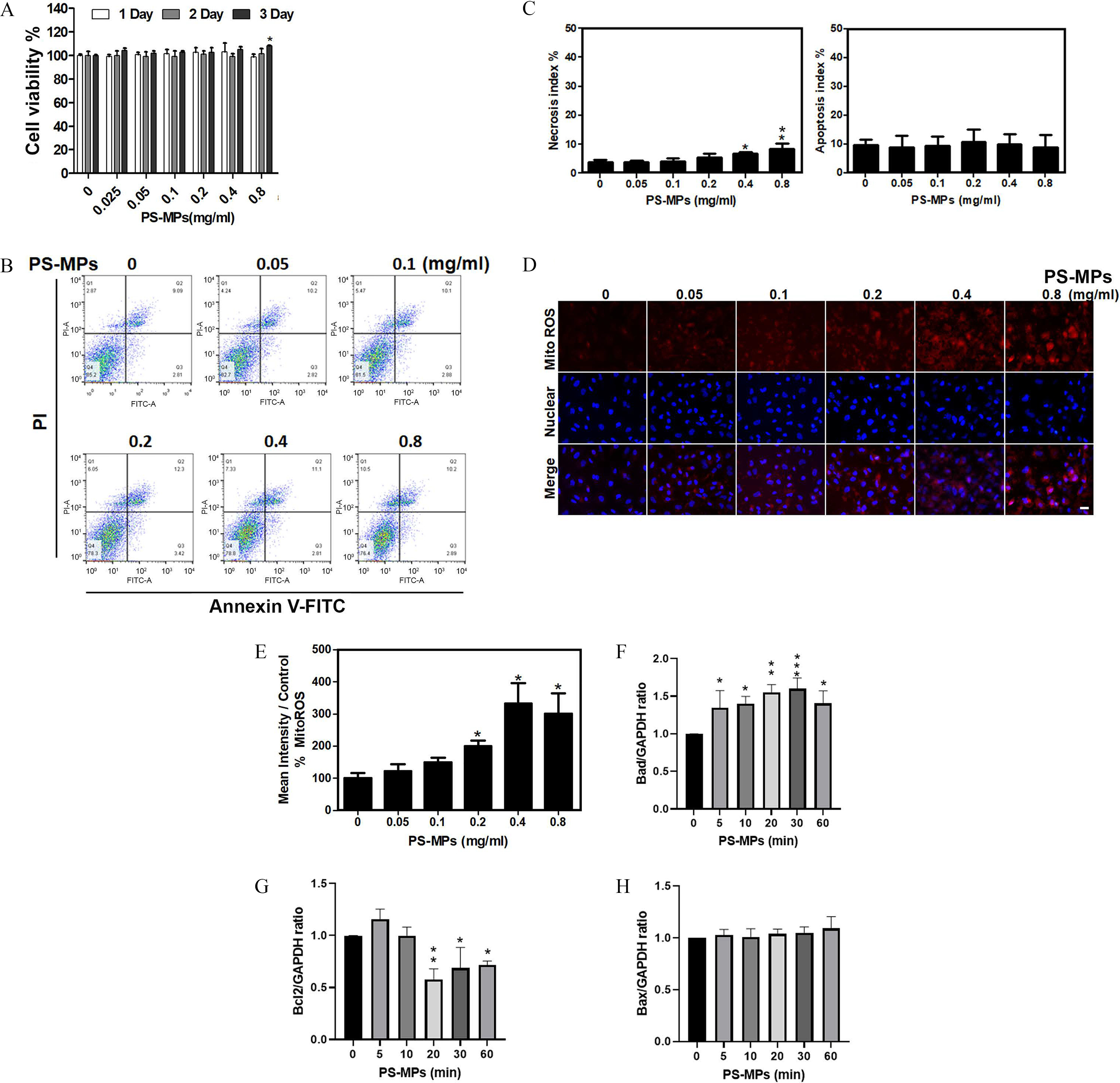
Cell viability, apoptosis, necrosis, mitochondrial ROS levels, and apoptosis-related proteins expression in HK-2 cells treated with PS-MPs. (A) Cell viability was determined after treatment with PS-MPs at concentrations of 0.05, 0.1, 0.2, 0.4, and for 1, 2, and 3 d. . * compared with the control group, as determined by two-way ANOVA with Dunnett’s multiple comparisons test. Significance: Day 3, group vs. group: . The mean and SD summary data of (A) are shown in Table S3. For all -values, see Table S4. (B) Apoptosis was detected by flow cytometry after treatment with PS-MPs at concentrations of 0.05, 0.1, 0.2, 0.4, and for 24 h. (C) The necrosis and apoptosis indexes were graphed and analyzed. . * and ** compared with the control group, as determined by one-way ANOVA with Dunnett’s multiple comparison test. Significance: necrosis index: group vs. group: , group vs. group: ; apoptosis index: no significant differences (Table S4). The mean and SD summary data of (C) are shown in Table S3. (D) Mitochondrial ROS were detected with MitoSOX Red after treatment with PS-MPs at concentrations of 0.05, 0.1, 0.2, 0.4, and for 6 h. DAPI (blue) was used for nuclear staining. . (E) Mitochondrial ROS levels were graphed and statistically analyzed. . * and *** compared with the control group, as determined by one-way ANOVA with Dunnett’s multiple comparison test. Significance: group vs. group: , group vs. group: , group vs. group: . The mean and SD summary data of (E) are shown in Tables S3 and S4. The levels of apoptosis-related proteins, such as (F) Bad, (G) Bcl2, and (H) Bax, were assessed after treatment with PS-MPs at a concentration of for 0, 5, 10, 20, 30, and 60 min. GAPDH served as an internal control. The data are presented as the . The mean and SD summary data for quantification of western blots are shown in Table S3. The western blotting results were quantified and statistically analyzed as shown in Figure 2. Note: ANOVA, analysis of variance; DAPI, 4,6-dimidyl-2-phenylindole; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HK-2 cells, human kidney 2 cells; PS-MPs, polystyrene microplastics; ROS, reactive oxygen species; SD, standard deviation.
