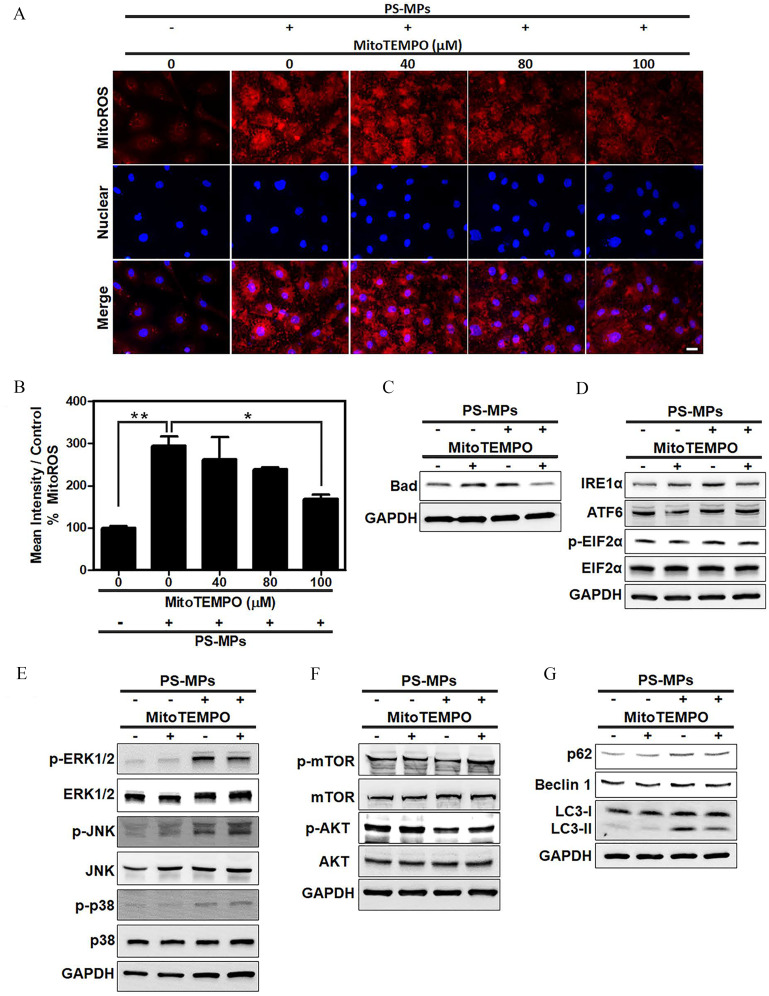
Figure 4.
Levels of Bad, ER stress-related proteins, MAPK-, and AKT/mTOR signaling pathway proteins and autophagy-related proteins after PS-MP treatment in HK-2 cells with mitochondrial ROS inhibition. (A) Mitochondrial ROS levels (red) using MitoSOX Red stain after pretreatment with MitoTEMPO at concentrations of 40, 80, and . After MitoTEMPO pretreatment, PS-MPs were added at a concentration of . DAPI (blue) was used for nuclear staining. . (B) The mean intensity after inhibition of mitochondrial ROS production was normalized to that in the control group and graphed. . * and ** compared with the control group, as determined by one-way ANOVA with Dunnett’s multiple comparison test. Significance: PS-MPs group vs. 0 PS-MPs mg/mL group: , PS-MP–only group vs. MitoTEMPO group: . The mean and SD summary data of (B) are shown in Table S3. See Table S4 for -values for all comparisons. (C) The expression of apoptosis-related protein Bad was examined. Cells were pretreated with MitoTEMPO () for 1 h, PS-MPs () were added, and the cells were incubated for another 20 min. (D) The expression of ER stress-related proteins, such as , ATF6, , and , was examined. Cells were pretreated with MitoTEMPO for 1 h, PS-MPs were added, and the cells were incubated for another 24 h. (E) The phosphorylation of MAPK signaling pathway components p38, ERK1/2, and JNK was examined. Cells were pretreated with MitoTEMPO for 12 h, PS-MPs were added, and the cells were incubated for another 30 min. (F) The phosphorylation of AKT/mTOR pathway components mTOR, and AKT was examined. Cells were pretreated with MitoTEMPO for 1 h, PS-MPs were added, and the cells were incubated for another 1 h. (G) The expression of autophagy-related proteins p62, Beclin 1, and LC3 was examined after pretreatment with MitoTEMPO for 1 h and treatment with PS-MPs for 24 h. GAPDH served as an internal control. The data are presented as the . The mean and SD summary data for quantification of western blots are shown in Table S3. The western blotting results were quantified and statistically analyzed, as shown in Figure S8. Note: AKT, protein kinase B; ANOVA, analysis of variance; ATF6, activating transcription factor 6; DAPI, 4,6-dimidyl-2-phenylindole; , eukaryotic initiation factor 2 alpha; ER, endoplasmic reticulum; ERK1/2, extracellular signal-regulated kinases 1 and 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HK-2 cells, human kidney 2 cells; , inositol-requiring enzyme ; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; mTOR, mitogen-activated protein kinase; p, phosphorylated; PS-MPs, polystyrene microplastics; ROS, reactive oxygen species; SD, standard deviation.