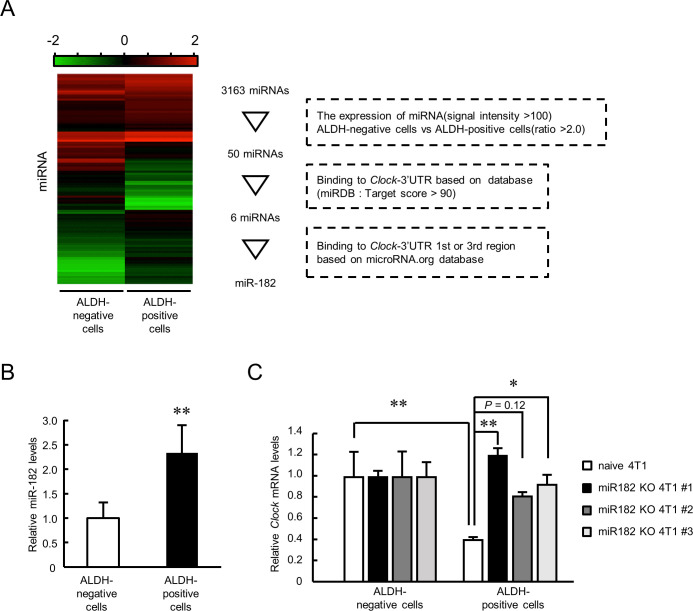
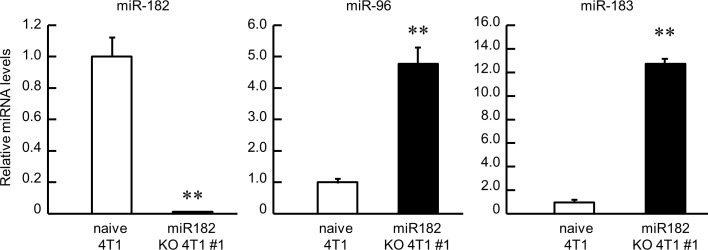
Figure 6. Role of miR-182 in post-transcriptional regulation of Clock gene expression in ALDH-positive 4T1 cells.
(A) Procedure of selecting miRNA that regulates Clock expression in ALDH-positive 4T1 cells. Heatmap shows the differential expression of miRNA between ALDH-negative and -positive 4T1 cells. microRNA target prediction databases (miRDB and microRNA.org) were applied to this selection. (B) The expression levels of mmu-miR-182 in ALDH-negative and -positive cells. Data were normalized by the β-Actin mRNA levels. Values are the mean with SD (n = 5). The value of ALDH-negative cells is set at 1.0. **p<0.01; significant difference from ALDH-negative cells (t8 = 3.828, Student’s t-test). (C) miR-182 negatively regulates the expression of Clock mRNA in ALDH-positive 4T1 cells. Three miR-182 KO clones were selected for this experiment. Data were normalized by the 18 s rRNA levels. Values are the mean with SD (n = 3). The values of ALDH-negative cells are set at 1.0. *p<0.05, **p<0.01; significant difference between two groups (F7,16 = 7.681, p<0.01, ANOVA with the Tukey–Kramer post hoc test).