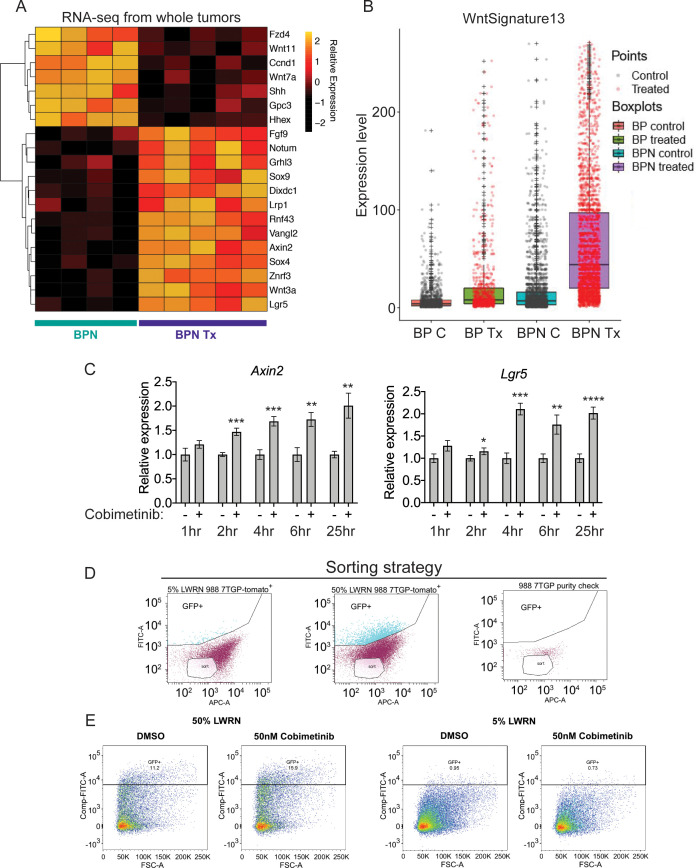
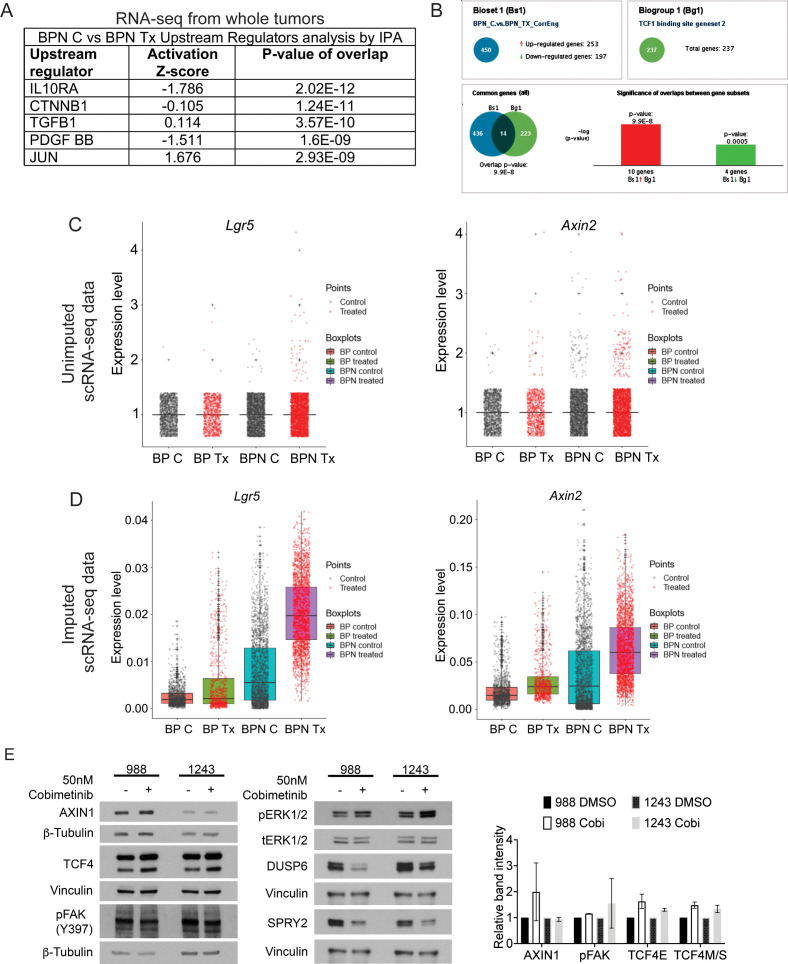
Figure 7. MAPK inhibition activates WNT signaling in NKX2-1-negative tumors.
(A) Transcriptome analysis of genes comprising the canonical WNT pathway gene ontology (AmiGO) in RNA purified from FACS-sorted BPN tumor cells 1 week post- treatment with PLX4720+PD0325901 or control chow. adjP <0.05 for each comparison. Color key indicates normalized expression levels (Log10). (B) Analysis of WNT pathway activity in scRNA-seq data using a WNT activation signature. BP indicates cells from clusters 4–8; BPN indicates cells from clusters 1–3 (see Figure 4E, F). C : control, Tx = drug treatment. (C) Kinetics of WNT signaling induction following MAPK-inhibitor treatment as indicated by qRT-PCR analysis of the canonical WNT signaling markers, Axin2 and Lgr5, on RNA isolated from drug/DMSO treated organoids for the indicated times. Data are mean ± S.D. A representative experiment of three is shown. ****p<0.0001, ***p<0.001, **p<0.01, *p<0.05 by Student’s t-test. (D, E) WNT-low cells can give rise to WNT-high cells. BPN organoid lines, 988 shown here, were stably transduced with a WNT-reporter construct (7TGP, Addgene Plasmid #24305) and FACS-sorted to isolate cells with undetectable WNT activity (pink gate) (D). (E) Dynamics of sorted WNT-low cells when cultured in WNT rich spheroid media (50% LWRN vs. 5% LWRN) or in the presence of MEK-inhibitor. Numbers shown are the percentage of live tdTomato+GFP+ cells in each culture as quantified by flow cytometry (FlowJo). Data shown is a representative of two independent sorting experiments.