Fig. 1.
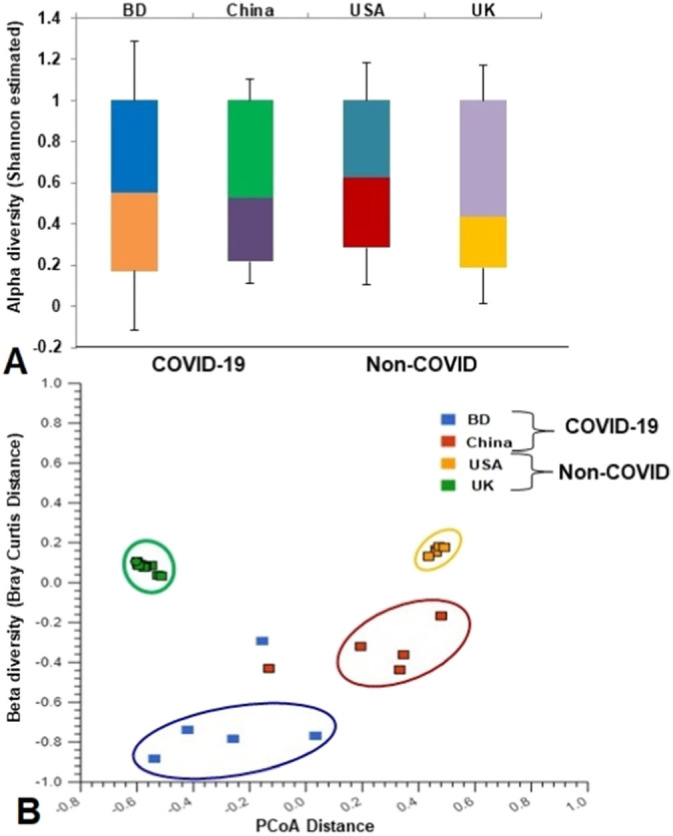
Differences in microbiome diversity and community structure in COVID-19 (BD and China), and non-COVID (UK and USA) disease metagenomes. (A) Box plots showing significant differences (P = 0.036, Kruskal-Wallis test) in Shannon estimated alpha diversity in four metagenomes. (B) Principal coordinates analysis (PCoA) measured on the Bray-Curtis distance method separated samples by microbial population structure. Each dot represents an individual, and colors indicate the populations in four metagenomes. Statistical analysis using Kruskal–Wallis tests showed significant microbial diversity variations across the four metagenomes (P = 0.021, Kruskal-Wallis test).
