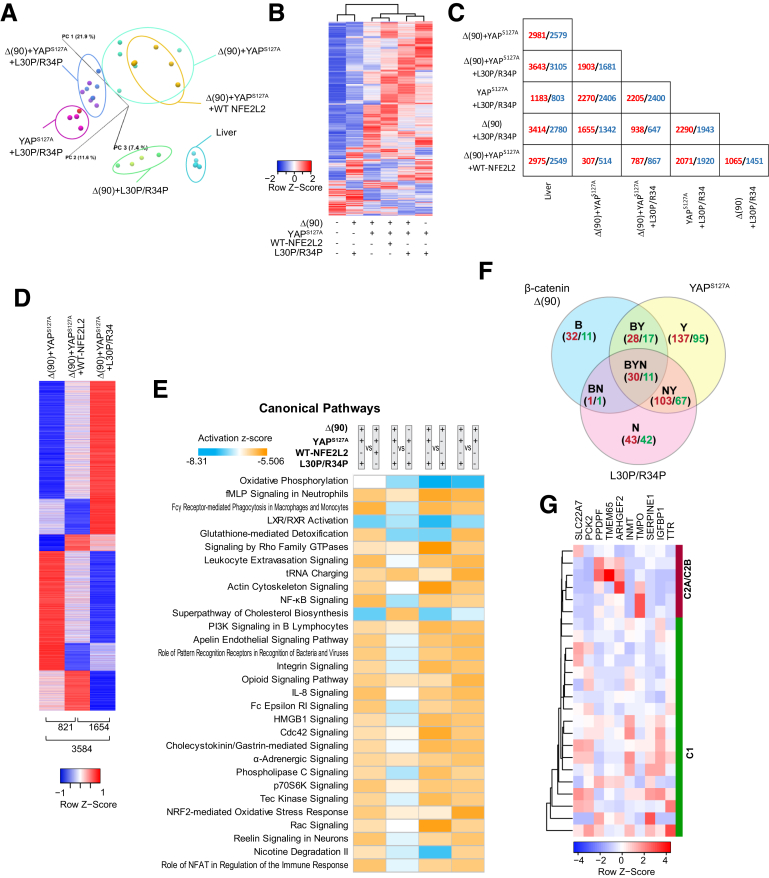
Figure 5.
RNAseq analysis of tumors generated by combinations of Δ(90), YAPS127A, WT-NFE2L2, L30P, and R34P. (A) Principal components analysis of transcriptomic profiles of livers and tumors (n = 4–5 samples/group. (B) Heat maps of differentially expressed transcripts from the tissues depicted in (A) arranged by hierarchical clustering. Because only a single transcript difference was found between tumors expressing L30P and R34P, results were combined for this and subsequent analyses (L30P/R34P). (C) Pairwise comparisons showing the number of significant gene expression differences between any 2 of the tissues depicted in (B). Red and blue, up-regulated and down-regulated, respectively, in the tumors depicted at the left relative to those indicated at the bottom. (D) Distinct transcript patterns of Δ(90)+YAPS127A, Δ(90)+YAPS127A+WT-NFE2L2, and Δ(90)+YAPS127A+L30P/R34P tumor cohorts. Numbers at bottom of each column indicate the significant expression differences between each pairwise comparison. (E) Top IPA pathways among different tumor groups, expressed as z-scores. Orange, up-regulated; blue, down-regulated. (F) Shared gene expression subsets between and among the indicated cohorts. Red and green, number of transcripts up-regulated and down-regulated, respectively, relative to liver. (G) Hierarchical clustering of C1 and C2A/C2B subsets31,32 of human HBs using the 10 “BYN” transcripts from Table 2 that were dysregulated in human tumors.