Extended Data Fig. 7. Flow cytometry analysis of scRNAseq-identified human WAT macrophage and dendritic cell populations.
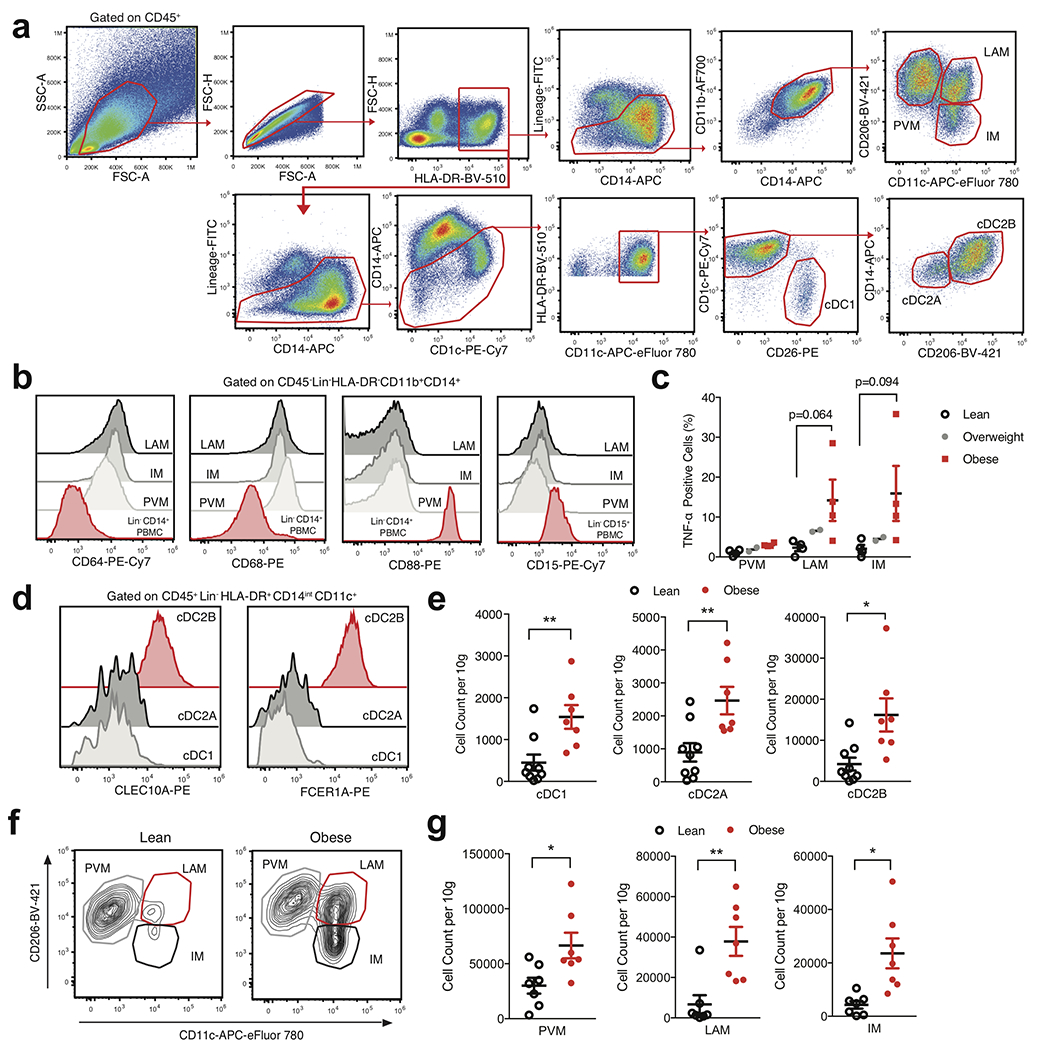
(a) Representative gating strategy for scRNAseq-defined human WAT macrophage and DC populations identified in Figure 4. (b) Representative histograms of CD64, CD68, CD88, and CD15 expression on human WAT macrophage cell subsets, CD14+ monocytes and CD15+ neutrophils isolated from human PBMC. (c) Flow cytometry analysis of endogenous TNF-α production by human WAT macrophage subsets from n=4 lean, n=2 overweight, and n=4 obese patients. Each point represents an individual patient. (d) Representative flow cytometry histograms of CLEC10A and FCER1A expression on human WAT DC subsets. (e) Density of the indicated DC populations by BMI classification, n=9 lean and n=7 obese patients. Each point represents an individual patient (f) Representative flow cytometry plots of human WAT macrophage populations from lean (left) and obese (right) patients. (g) Density of the indicated macrophage populations by BMI classification, n=7 lean and n=7 obese patients. Each point represents an individual patient. (b,d) Data is representative of 3 individual patient samples. Samples were compared using two-tailed Student’s t test with Welch’s correction, assuming unequal SD, and data are presented as individual points with the mean ± SEM (*p<0.05, **p<0.01).
