Figure 7. CellphoneDB analysis predicts a dramatic remodeling of the human WAT interactome during obesity.
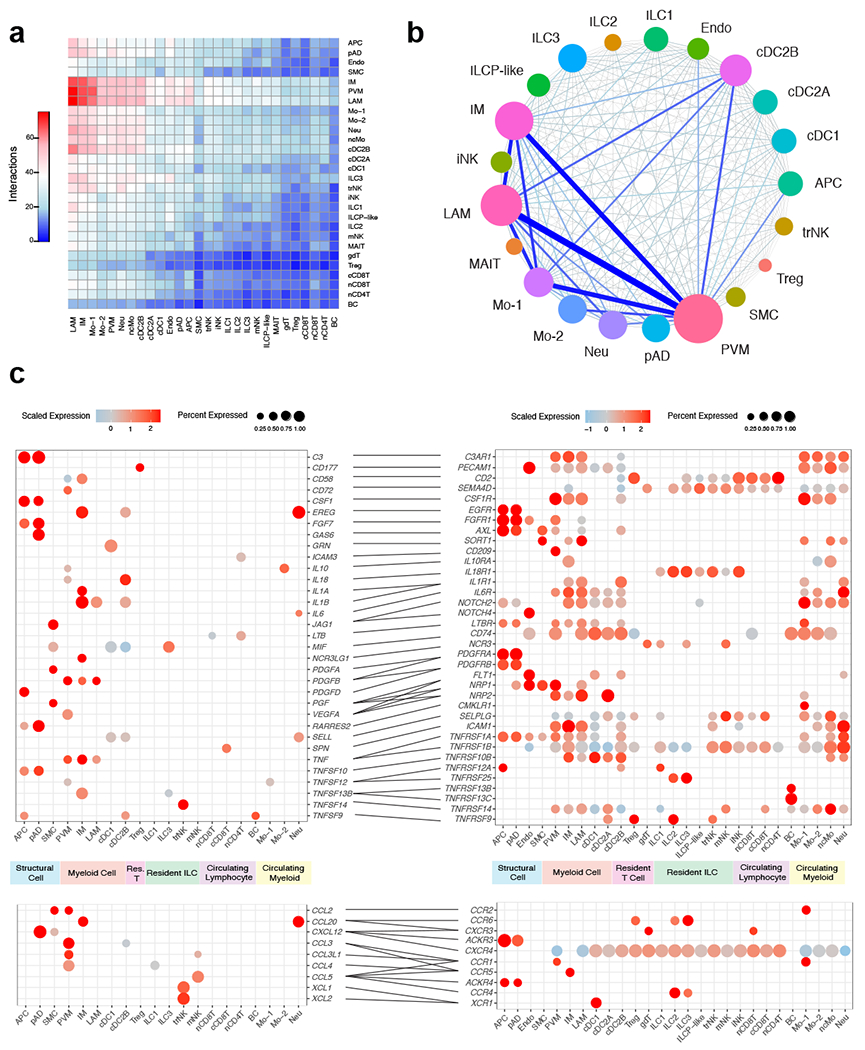
(a) Interaction heatmap plotting the total number of lean WAT-derived cell receptor (y-axis) and ligand (y-axis) interactions for the specified cell types. Color represents the number of interactions between cell types; higher number of interactions (red), lower number of interactions (blue). (b) Connectome web analysis of obese highly interacting cell types, based on expression of the ligand in at least 25% of the cell population. Vertex (colored cell node) size is proportional to the number of interactions to and from that cell, while the thickness of the connecting lines is proportional to the number of interactions between two nodes. (c) Dot plots showing expression of ligands (left) and receptors (right) in human WAT cells; only ligands and receptors from cell types with detected expression (>25%) are shown. Implicated chemokines can be found in the lower panel. Color saturation indicates the strength of expression in positive cells, while dot size reflects the percentage of each cell cluster expressing the gene.
