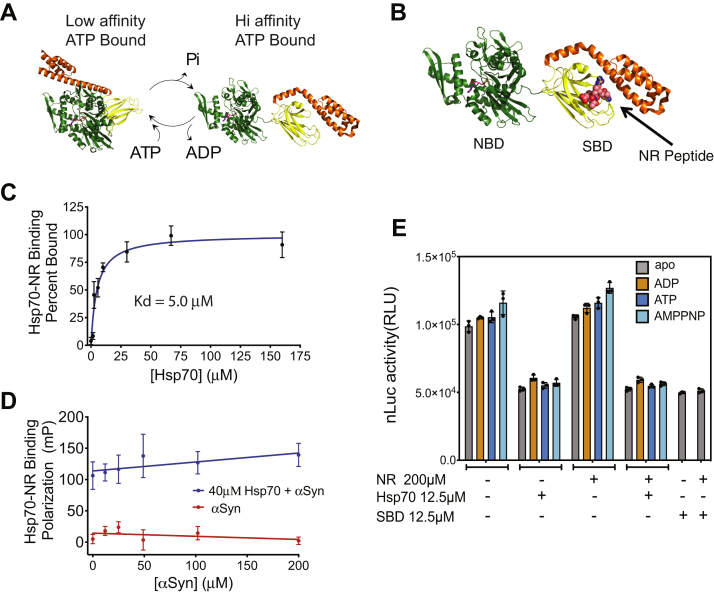
Figure 4.
Hsp70 blockage of ASyn oligomerization requires neither ATP cycling nor binding to the canonical substrate-binding site.A, schematic of canonical action of Hsp70 family members. Structures are based on the E. Coli Hsp70 homologue, DnaK. ATP hydrolysis drives Hsp70 from a low- (ATP) to a high-affinity (ADP) substrate-binding state. Nucleotides bind to the N-terminal nucleotide-binding domain (NBD, green, residues 1–388). Substrate binds to a pocket formed in the C-terminal substrate-binding domain (SBD, yellow/orange residues (389–602) by the lid region (orange, residues 508–602) folding over the β-sheet core of the SBD (yellow, residues 389–507) in the ADP-bound conformation. Protein data bank identifiers are 2kho, 4b9q, and 1dkx. B, structure of NR peptide bound to the E. coli Hsp70 homologue DnaK is shown. NR peptide binds to the canonical substrate-binding site. C, binding of NR peptide to Hsp70 was measured by fluorescence polarization of N-terminal 5-FAM (5-Carboxyfluorescein) label on the NR peptide as described in methods. NR binds with a Kd of 5.0 μM. Means ± s.d. shown, n = 6. D, a competitive binding assay shows that the binding of NR peptide to Hsp70 is not competed by the presence of increasing ASyn (αSyn-WT). Means ± s.d. shown, n = 6. E, 10 μM each of ASyn-LgBiT and ASyn-SmBiT, with or without full-length Hsp70 or Hsp70-SBD (see Fig. 5), 2 mM nucleotide and the NR peptide as indicated were incubated in 384-well plates for 24 h and nLuc luciferase activity measured as described in methods. ADP stabilizes the substrate-bound conformation, and AMPNP, a nonhydrolyzable ATP analogue, stabilizes the ATP-bound state. Neither nucleotide, NR nor truncation impacted Hsp70 blockage of ASyn oligomerization. Means ± s.d. shown, n = 3. Lower concentrations of NR peptide likewise had no impact on Hsp70 blockage of ASyn oligomerization (Fig. S6).