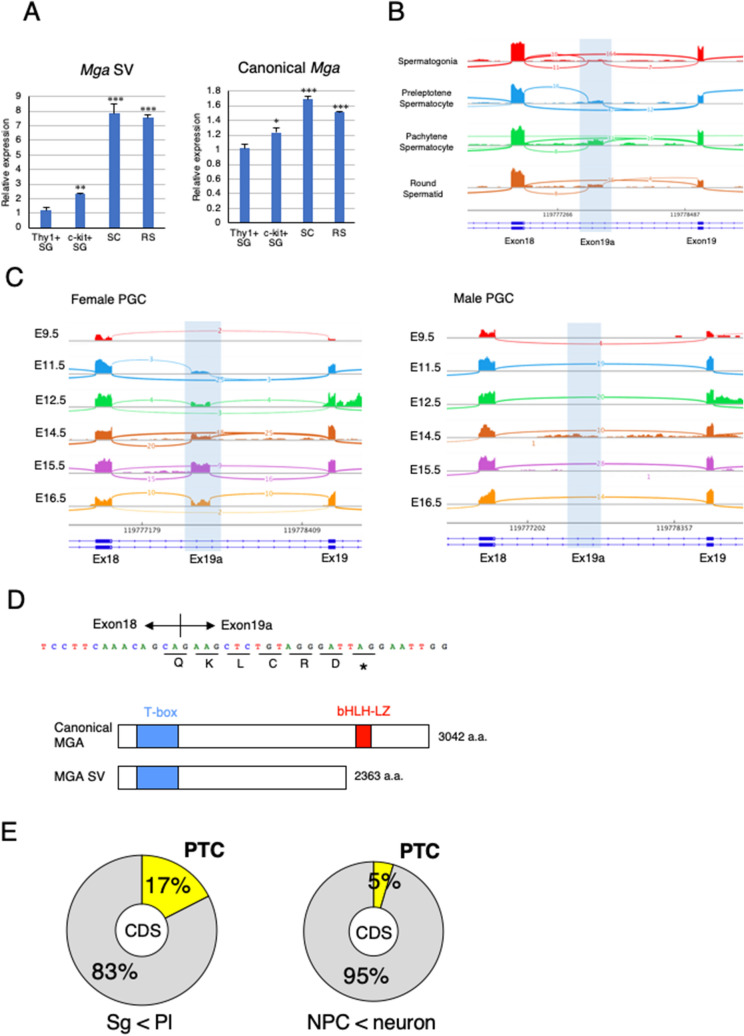
Figure 3.
Mga splice variant is restrictively present around the meiotic stage of germ cells. (A) qPCR analyses of RNAs from undifferentiated (Thy1+) and differentiated (c-Kit+) spermatogonia (SG), spermatocytes (SC), and round spermatids (RS) to determine the levels of Mga SV (left panel) and canonical Mga (right panel) mRNAs. Values obtained from Thy1+ spermatogonia were arbitrarily set to one for both Mga mRNA species. Data were subjected to statistical analyses as described in Fig. 1B. **P < 0.01; ***P < 0.001 (B) Visualization of splicing events around exons 18 and 19 of the Mga gene during spermatogenesis in the testis by Sashimi plots. Plots were constructed using publicly reported data (GSE75826). Abbreviations are described in (A). (C) Sashimi plots showing splicing events around exons 18 and 19 of the Mga gene in male and female PGCs during the embryonic stage. Data for female and male PGCs were obtained from publicly reported data (E-MTAB-4616) and shown in left and right panels, respectively. (D) Sequence of exon 19a containing the in-frame PTC. Acquisition of the exon 19a sequence by alternative splicing was accompanied by insertion of PTC after the addition of the coding sequence of five amino acids, leading to translation into the carboxy-terminally truncated anomalous MGA protein that lacked the bHLHZ domain as depicted in lower portion. (E) Generation of the PTC-containing transcript by alternative splicing was a frequent event during meiotic onset of germ cells. Frequencies of exon inclusion leading to incorporation of a PTC and that leading to addition of the novel amino acid sequence occurred during conversion of spermatogonia to meiotic germ cells (left) and differentiation of neural progenitor cells into neuronal cells (right) are presented as pie charts. Publicly reported RNA sequence data of germ cells (GSE75826) and neuronal cells (GSE96950) were used to calculate the frequencies.