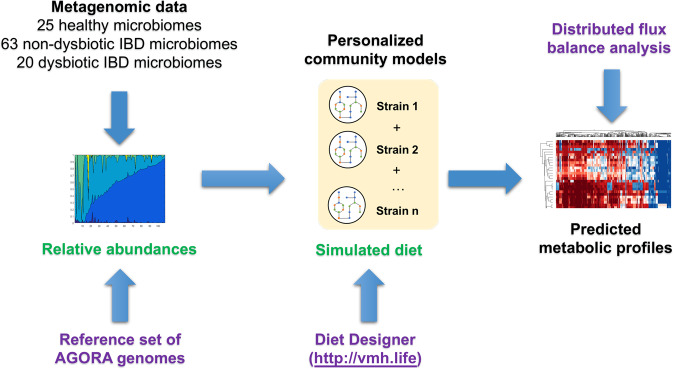
Fig. 1. Schematic overview of the modelling framework established in this study.
Metagenomic reads were mapped onto a reference set of AGORA18 genomes, and the strain-level relative abundances were retrieved. For each metagenomic sample, a personalised microbiome model was constructed. The models were parameterised further with a simulated “Average European” diet retrieved from the Virtual Metabolic Human17 database. The metabolic profile of each microbiome model was then computed with distributed flux balance analysis53. Required input data are shown in green and required tools are shown in purple.