Abstract
Abnormally increased resorption contributes to bone degenerative diseases such as Paget’s disease of bone (PDB) through unclear mechanisms. Recently, the optineurin (OPTN) gene has been implicated in PDB, and global OPTN knockout mice (Optn−/−) were shown to exhibit increased formation of osteoclasts (osteoclastogenesis). Growing evidence, including our own, has demonstrated that intracellular reactive oxygen species (ROS) stimulated by receptor activator of nuclear factor kappa-B ligand (RANKL) can act as signaling molecules to promote osteoclastogenesis. Here, we report that OPTN interacts with nuclear factor erythroid-derived factor 2-related factor 2 (NRF2), the master regulator of the antioxidant response, defining a pathway through which RANKL-induced ROS could be regulated for osteoclastogenesis. In this study, monocytes from Optn−/− and wild-type (Optn+/+) mice were utilized to differentiate into osteoclasts, and both qRT-PCR and tartrate-resistant acid phosphatase (TRAP) staining showed that the Optn−/− monocytes exhibited enhanced osteoclastogenesis compared to the Optn+/+ cells. CellROX® staining, qRT-PCR, and Western blotting indicated that OPTN deficiency reduced the basal expression of Nrf2, inhibited the expression of NRF2-responsive antioxidants, and increased basal and RANKL-induced intracellular ROS levels, leading to enhanced osteoclastogenesis. Coimmunoprecipitation (co-IP) showed direct interaction, and immunofluorescence staining showed perinuclear colocalization of the OPTN-NRF2 granular structures during differentiation. Finally, curcumin and the other NRF2 activators attenuated the hyperactive osteoclastogenesis induced by OPTN deficiency. Collectively, our findings reveal a novel OPTN-mediated mechanism for regulating the NRF2-mediated antioxidant response in osteoclasts and extend the therapeutic potential of OPTN in the aging process resulting from ROS-triggered oxidative stress, which is associated with PDB and many other degenerative diseases.
Subject terms: Mechanisms of disease, Stress signalling
Bone degeneration: Identifying a damage-limiting protein
The protein optineurin is implicated in regulating the breakdown of bone during natural maintenance by a mechanism that may be relevant to understanding and treating degenerative bone diseases. Cells called osteoclasts mediate bone breakdown during natural bone turnover, but excessive breakdown occurs in some bone diseases. Working with cultured mouse cells, researchers in the USA, led by Ching-Chang Ko at Ohio State University, Columbus, and Henry Tseng at Duke University, identified a molecular signaling pathway in which optineurin interacts with a protein called NRF2, which is known as the master regulator of the antioxidant response. This interaction allows optineurin to limit the production of chemicals called reactive oxidative species which stimulate osteoclast generation. The findings suggest that drugs regulating the optineurin signaling pathway may help treat bone degeneration and possibly other degenerative diseases.
Introduction
During skeletal remodeling, bone formation by osteoblasts is balanced with bone degradation by osteoclasts to meet the functional demands of the skeletal system1,2. An imbalance due to hyperactive osteoclastic activity results in net bone loss3,4 and abnormal remodeling, contributing to severe degenerative bone diseases such as Paget’s disease of bone (PDB), a chronic disorder most commonly affecting older people5–8. PDB is increasing in prevalence with the aging population and afflicts millions of individuals worldwide9,10. There is no cure or effective treatment for bone degeneration caused by hyperactivated osteoclastogenesis in PDB, and the underlying molecular mechanisms remain unknown.
Mature osteoclasts are derived from differentiating osteoclast precursors. This process, called osteoclastogenesis, is triggered by the binding of receptor activator of nuclear factor kappa-B ligand (RANKL) to its receptor on osteoclastic progenitors (preosteoclasts), which activate multiple downstream signaling pathways11, in which intracellular reactive oxidative species (ROS) have recently been demonstrated to serve as critical secondary messengers12–15. In contrast to physiological levels of ROS, which serve as signaling molecules16–18, excess ROS levels may result in abnormally elevated osteoclastogenesis, oxidative stress, and cellular damage over time19,20. Conversely, ROS scavengers have been consistently shown to inhibit osteoclastogenesis21–29. Beyond bone degeneration, ROS have also been broadly associated with age-dependent cellular degeneration in other tissues30–32.
Optineurin (OPTN), an adapter protein involved in numerous cellular functions, has recently been implicated in PDB33–35. We previously showed that aged mice genetically lacking optineurin (OPTN)36 exhibited hyperactivated osteoclastogenesis, focal bone lesions, and other PDB-like clinical features, while young mice did not display these phenotypes, although the mechanisms remain undefined. OPTN is expressed in bone and neuronal cells and has been reported to protect cells from ROS-induced cell damage37,38, suggesting a regulatory role of ROS in OPTN-mediated osteoclastogenesis. However, how OPTN regulates ROS homeostasis in osteoclasts remains unexplored.
Intracellularly, ROS homeostasis is mediated by nuclear factor-erythroid 2-related factor 2 (NRF2; also known as NFE2L2), a transcription factor that acts as a master regulator of the cellular antioxidant response. NRF2 has a basic-region leucine zipper (bZIP) domain39 that allows it to heterodimerize with other proteins, regulates its stability, and modulates transcriptional activity40,41. Interestingly, OPTN also contains a bZIP domain42, identifying it as a potential NRF2 binding partner that might contribute to the regulation of ROS levels and osteoclastogenesis43. However, the direct Nrf2 and OPTN interaction as a ROS homeostatic mechanism has not been demonstrated.
In this study, we tested the hypothesis that OPTN interacts with NRF2 to modulate RANKL-induced ROS signaling and osteoclastogenesis. Our results will provide novel insights into how loss of normal OPTN function results in attenuated antioxidant responses and elevated osteoclastogenesis. These insights will improve our understanding of abnormal bone remodeling and the development of therapies for bone diseases resulting from increased osteoclast activities. Because OPTN is also genetically associated with other degenerative diseases, such as glaucoma and amyotrophic lateral sclerosis (ALS), our study will potentially offer broader insights into the pathophysiology and treatment of various degenerative conditions.
Materials and methods
Chemicals
Receptor activator of nuclear factor kappa-B (NF-κB) ligand (RANKL, #462-TEC-010) and macrophage colony-stimulating factor (M-CSF, #416-ML-010) were obtained from R&D Systems (Minneapolis, MN). The RealTime-Glo™ MT Cell Viability Assay was purchased from Promega (Madison, WI). RNAzol was purchased from the Molecular Research Center (Cincinnati, OH). The iScript™ cDNA Synthesis Kit for RT-qPCR was purchased from Bio-Rad (Hercules, CA). The dye 2′,7′-dichlorofluorescin diacetate (DCF, #D6883) for ROS measurement was purchased from Sigma-Aldrich (St. Louis, MO). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS), penicillin, and streptomycin were all obtained from Thermo Fisher Scientific (Waltham, MA). The NRF2 inducers curcumin (CUR, #C1386), quercetin (QUE, #Q4951), bardoxolone methyl (CDDO-Me, #SMB00376), sulforaphane (SFN, #S4441), and tert-butylhydroquinone (tBHQ, #112941) were all purchased from Sigma-Aldrich.
Animals
Global Optn knockout (Optn−/−) mice on a C57BL/6 background were generated by crossing Optnflox/flox mice with CMV-Cre mice (Jackson Laboratories, Bar Harbor, ME) as described in our previous study36. All animal procedures were approved by the Institutional Animal Care and Use Committees at the University of North Carolina at Chapel Hill and Duke University.
Cell culture
For primary culture of osteoclast precursors, 8- to 12-week-old Optn−/− and Optn+/+ mice were euthanized by CO2, and tibias and femurs were collected. Bone marrow cells were flushed out into α-MEM medium containing 10% FBS and penicillin/streptomycin and cultured at 37°C in a humidified 5% CO2 atmosphere. After one day, nonadherent cells were collected and reseeded in plates with 30 ng/mL M-CSF for 3 days to grow osteoclast precursor cells. Cycloheximide (0.5 mM, C7698-1G, Sigma-Aldrich) was used to inhibit protein synthesis.
Osteoclast differentiation and TRAP staining
For osteoclast differentiation, osteoclast precursors in 48-well plates were treated with RANKL (10 ng/mL) and M-CSF (30 ng/mL) for 6 days. The cells were fixed with 10% paraformaldehyde for 10 min before incubation in TRAP staining solution for 10–30 min to detect TRAP+ multinucleated osteoclast-like cells. TRAP-stained plates were scanned and imaged using an Eclipse Ti microscope (Nikon, Shinagawa, Japan).
Cell viability assay
Cell viability was measured by the RealTime-Glo™ MT Cell Viability Assay according to the manufacturer’s directions (Promega). In brief, a cell suspension was prepared by adding MT Cell Viability reagents to the culture media. Cells were seeded in this medium and cultured for up to 3 days. The luminescence of the cells was measured via a Cytation 5 plate reader (BioTek, Winooski, VT). Cell viability is expressed as the relative fold change compared to that of control cells cultured in normal growth medium.
Measurement of intracellular ROS
Qualitative and quantitative analyses of intracellular ROS in compound-treated cells were performed by fluorescence microscopy using CellROX Green/Nucblue staining reagents. ROS levels were also assessed by a plate reader using 2′,7′-dichlorofluorescin diacetate (DCF) as previously described29. In brief, osteoclast precursors were incubated with 10 μM DCF for 15 min at 37°C, rinsed in PBS three times, stimulated with RANKL (100 ng/mL) for up to 15 min and lysed with Tris-HCl (0.01 M)/0.5% Triton X-100 (pH 7.4). Fluorescence of the lysate was measured by a plate reader at 485 nm excitation and 535 nm emission.
RT-qPCR
Total mRNA of osteoclast precursor cells was extracted using RNAzol and reverse-transcribed using the iScript™ Kit. Subsequently, PCR reactions were prepared by using iTaqTM Universal SYBR Green Supermix and performed on a StepOnePlus Real-time PCR system (Applied Biosystems). Primers for target genes are listed in Supplementary Table S1. Threshold cycles of primers were normalized to the housekeeping gene β-actin, and the relative values were calculated based on the comparative Ct method (2−ΔΔCt method) as previously described29.
Western blot
Protein samples were prepared either by RIPA buffer (R0278, Sigma-Aldrich) to yield total protein or by a nuclear extraction kit (Cat. #2900, Millipore Sigma) to yield cytoplasmic and nuclear proteins. The Criterion Vertical Electrophoresis Cell and Trans-Blot Turbo Transfer System (Bio-Rad) was used for immunoblot analysis as described in our previous study29. Protein expression was detected by ECL Prime Western Blotting Detection Reagent (GE Healthcare Amersham). Primary antibodies used in the study were against NRF2 (#1272 T, Cell Signaling Technology), p-NRF2 Ser40 (#PA5-67520, Invitrogen), KEAP1 (#sc-514914, Santa Cruz Biotechnology), HMOX1 (#SC-136960, Santa Cruz Biotechnology), NQO1 (#sc-32793, Santa Cruz Biotechnology) and β-ACTIN (#SC-47778, Santa Cruz Biotechnology). HRP-linked anti-rabbit or anti-mouse IgG antibodies (#7074P2, #7076P2, Cell Signaling Technology) were used as secondary antibodies.
In vitro coimmunoprecipitation assays
The NC16 pCDNA3.1 FLAG NRF2 plasmid and pDEST26-OPTN plasmid were purchased from Addgene (Watertown, MA). HEK293T cells were seeded in 6-well plates and transfected with the indicated plasmids at a concentration of 2000 ng per plasmid/well by Lipofectamine 2000 according to the manufacturer’s protocol. Transfected cells were lysed in NP-40 buffer and split into two groups: 200 µl fractions for the input assay and 800 µl fractions for coimmunoprecipitation. The input lysates were resolved on SDS-PAGE gels and analyzed by immunoblot. The coimmunoprecipitated lysates were incubated with 20 µl of Anti-Flag® M2 Affinity Gel (Millipore Sigma A2220) for 24 h. After incubation, the samples were centrifuged at 5000 × g for 1 min, and the pellets were washed five times with NP-40 buffer. The proteins were finally resolved on SDS-PAGE gels and analyzed by immunoblot.
Confocal microscopy
Optn−/− and Optn+/+ preosteoclasts were fixed with 4% paraformaldehyde for 24 h. After three washes with PBS, the cells were permeabilized with 0.3% Triton X-100 for 10 min and washed three times again with PBS. After the cells were blocked with horse serum for 24 h, they were incubated with anti-rabbit OPTN (Abcam, ab23666, 1:150) and anti-rat NRF2 (Cell Signaling, #14596, 1:150) primary antibodies; anti-rat NRF2 (Cell Signaling, #14596, 1:150) and anti-mouse KEAP1 (Santa Cruz Biotechnology, #sc-514914, 1:50) primary antibodies; anti-mouse KEAP1 (Santa Cruz Biotechnology, #sc-514914, 1:50) and anti-rabbit SQSTM1/p62 (Abcam, ab240635, 1:150) primary antibodies overnight at 4°C. The cells were then incubated with goat anti-rabbit Alexa Fluor 488 (Thermo Fisher, #A32731, 1:200), goat anti-rat Alexa Fluor 594 secondary antibodies (Thermo Fisher, #A11007, 1:200), goat anti-mouse Alexa Fluor 488 (Thermo Fisher, #A28175, 1:200), and goat anti-mouse Alexa Fluor 594 (Thermo Fisher, #A11032, 1:200) accordingly for 2 h at room temperature. After DAPI staining, the cells were imaged by confocal microscopy using the FITC, CY3, and DAPI channels.
Image quantification
For protein quantification by western blotting, ImageJ (version 1.52k; Bethesda, MD) was used to analyze the gray value of each band to represent intensity, which was normalized to β-actin. For intracellular ROS quantification, ImageJ was applied to measure the area integrated intensity of the ROS fluorescence signal in each cell, and a total of 15 cells were randomly contoured and calculated in each image. The mean value of ROS fluorescence represented the intensity of each image. Each analysis was performed three times to determine statistical significance. For quantification of translocated NRF2, ImageJ was applied to identify the NRF2±DAPI± area (region of interest (ROI)) shown in white (Supplementary Fig. S2c). The mean fluorescence value of the ROI in each nucleus represents the intensity of translocated NRF2 per nucleus in each image. Each analysis was performed three times to determine statistical significance.
Statistical analyses
Statistical analyses were performed using Prism 5 (GraphPad Software, San Diego, CA). Significance was determined as p < 0.05. Comparisons with a specific control were assessed using one-way analysis of variance (ANOVA) followed by the Bonferroni t-test. Data are expressed as the mean ± standard error of the mean (SEM).
Results
OPTN deficiency enhances osteoclastogenesis
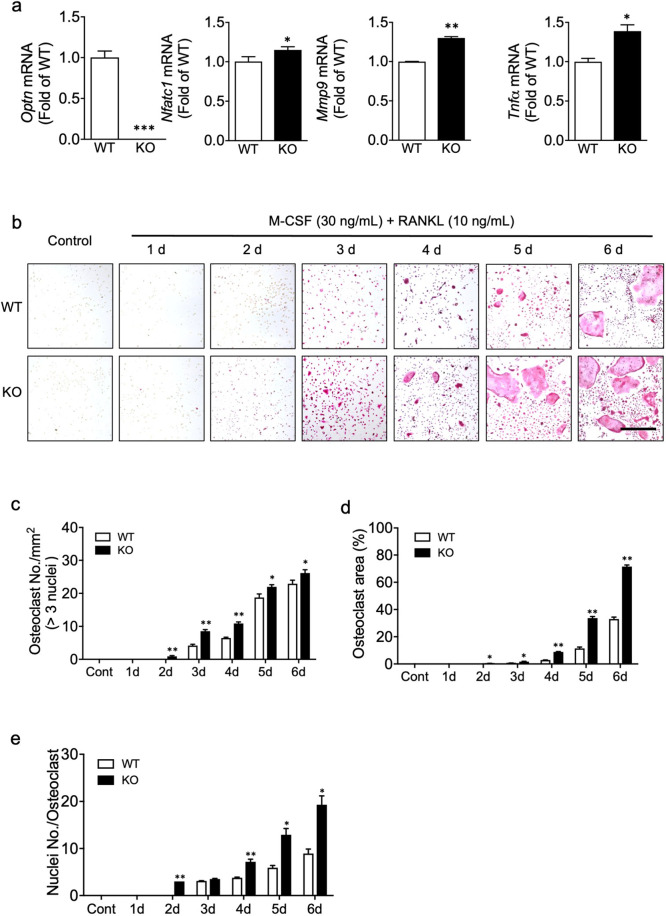
To test the effects of OPTN ablation on osteoclastogenesis, we determined the gene expression of Optn and osteoclast markers in osteoclast precursors under basal conditions (macrophages treated with M-CSF only). As expected, Optn transcripts were undetectable in the Optn−/− vs. Optn+/+ osteoclast precursors (Fig. 1a). In addition, the expression of several key osteoclastogenic markers, including Nfatc1, Mmp9, and Tnfα, was significantly increased in the Optn−/− vs. Optn+/+ macrophages, supporting a role for OPTN in osteoclast differentiation.
Fig. 1. OPTN deficiency enhances osteoclastogenesis.
a Higher expression of key osteoclast marker genes in OPTN KO vs. Optn+/+ preosteoclast precursors under basal, unstimulated conditions. b Formation of mature osteoclasts from precursors isolated from the Optn+/+ and Optn−/− mice. Primary osteoclast precursors were treated with RANKL (10 ng/mL) and M-CSF (30 ng/mL) for 6 days. Mature osteoclasts appear as multinucleated TRAP+ (red) cells. Representative images are shown in the panel. Scale bar = 500 μm. c Osteoclast number per mm2, d the percentage of osteoclast area per image, and e the number of nuclei per osteoclast. WT, Optn+/+; KO, Optn−/−; d, day. Control (growth media only). n = 3 experiments. Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 compared to Optn+/+ within each group.
To further investigate the role of OPTN, we treated the Optn+/+ and Optn−/− precursor cells with RANKL and M-CSF for 6 days to promote osteoclast differentiation, where precursors differentiate and merge into multinucleated giant TRAP+ cells that represent mature osteoclasts. Significantly enhanced osteoclastogenesis was observed in the Optn−/− vs. Optn+/+ cells (Fig. 1b), based on more mature osteoclasts per area (Fig. 1c), larger average osteoclast size (Fig. 1d), and more nuclei per cell (Fig. 1e). These results identify OPTN as a negative regulator of osteoclastogenesis.
OPTN deficiency results in elevated ROS production in preosteoclasts and osteoclasts
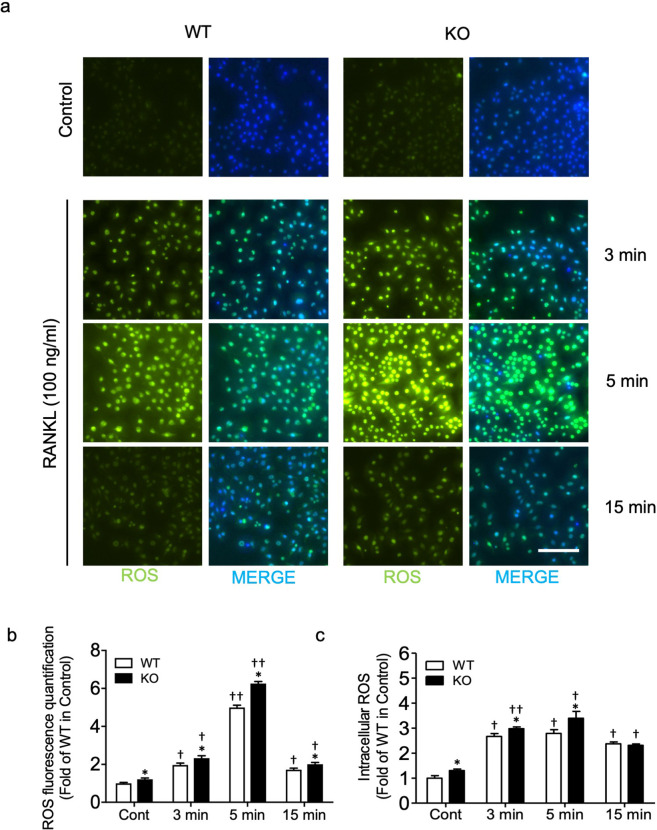
Among the various pathways that participate in osteoclastogenesis, ROS have been reported to act as secondary messengers during RANKL-induced osteoclast differentiation15,23,26,28,29,44. To further elucidate the role of ROS in OPTN-mediated osteoclastogenesis, we treated Optn+/+ and Optn−/− preosteoclasts with a high concentration of RANKL (100 ng/mL), which is reported to trigger downstream ROS signaling15,28,29, and assessed basal and RANKL-induced intracellular ROS levels by CellROX Green staining and fluorescence microscopy. In cells without RANKL treatment, we observed a higher level of basal ROS in the Optn−/− vs. Optn+/+ cells (Fig. 2a, first row). RANKL treatment induced significantly higher levels of ROS in both the Optn−/− and Optn+/+ cells than the control cells. Furthermore, the ROS level increased by 3 min, reached its peak at 5 min, and declined by 15 min (Fig. 2a, b), a pattern consistent with a previous report14. Since ROS are highly reactive and unstable, an additional quantitative analysis by DCF staining for intracellular ROS successfully confirmed these results (Fig. 2c). These findings revealed that loss of OPTN contributes to elevated intracellular ROS in both basal and RANKL-stimulated conditions.
Fig. 2. OPTN deficiency results in elevated ROS in preosteoclasts and osteoclasts.
a Merged images of CellROX Green fluorescence and DAPI staining in osteoclast precursors treated with RANKL (100 ng/mL) for the indicated time points (3, 5, and 15 min). Scale bar = 200 μm. b Quantification of CellROX Green fluorescence. c The intracellular ROS levels induced by RANKL (100 ng/mL) were determined using DCF staining and a plate reader. WT, Optn+/+; KO, OPTN-knockout; Cont, control (growth media only); n = 3 experiments. Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01 compared to Optn+/+ within each time point group; †p < 0.05, ††p < 0.01 compared to the untreated control of either the Optn+/+ or Optn−/− group.
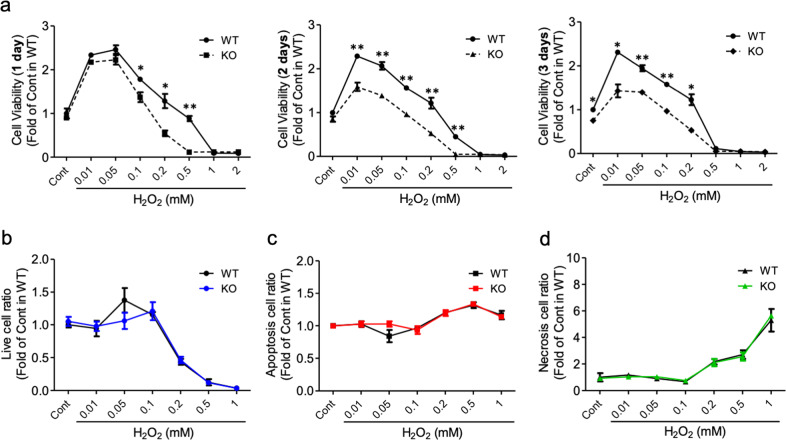
Optn−/− cells are more sensitive to H2O2-induced cytotoxicity than Optn+/+ cells
Notably, ROS levels in the Optn−/− cells were significantly higher than those in the Optn+/+ cells even under basal conditions, suggesting that the redox state in OPTN-deficient cells was affected (Fig. 2a, b). To further examine the antioxidant response in the Optn−/− cells, we treated these cells with H2O2, an oxidative stressor that promotes cellular oxidative damage. Using the MT cell viability assay to assess cell viability, we observed that both the Optn+/+ and Optn−/− cells were sensitive to H2O2 in a concentration-dependent manner. However, within the range of 0.01 to 1 mM, the Optn−/− cells were more susceptible to H2O2 than the Optn+/+ cells (Fig. 3a). Beyond 1 mM H2O2 exposure, cells with both genotypes showed complete cell death. In a time-course study, the Optn−/− cells were more sensitive than the Optn+/+cells to H2O2 exposure at 1, 2, and 3 days (Fig. 3a). To evaluate whether H2O2 caused apoptosis or necrosis in cells, we performed annexin-PI staining (Fig. 3b, c, d). Significant cell death began after 0.1 mM H2O2 treatment, consistent with the cell viability results. Furthermore, cell death was necrotic and not apoptotic for both genotypes, and no significant difference was observed between the two groups of cells for necrotic cell death. Taken together, our results showed that OPTN deficiency in preosteoclasts could increase cellular sensitivity to H2O2-induced cytotoxicity but primarily affects cell viability and not cell death.
Fig. 3. Optn−/− cells are more sensitive to H2O2-induced cytotoxicity.
a Cell viability of osteoclast precursors in response to various concentrations of H2O2. Each graph indicates a different treatment duration (1, 2, and 3 days). Quantitation of live b, apoptotic c, and necrotic d cells following treatment with various concentrations of H2O2. The cells are shown as a ratio of treated versus control untreated cells. The cells were treated with H2O2 for 1 day and then stained with Annexin V for apoptosis and necrosis before quantitation by flow cytometry. WT, Optn+/+ preosteoclasts; KO, Optn−/− preosteoclasts; Cont, control (growth media only); n = 3 experiments. Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01 compared to Optn+/+ within each dosage group.
Deficiency of OPTN attenuates Nrf2 and antioxidant expression
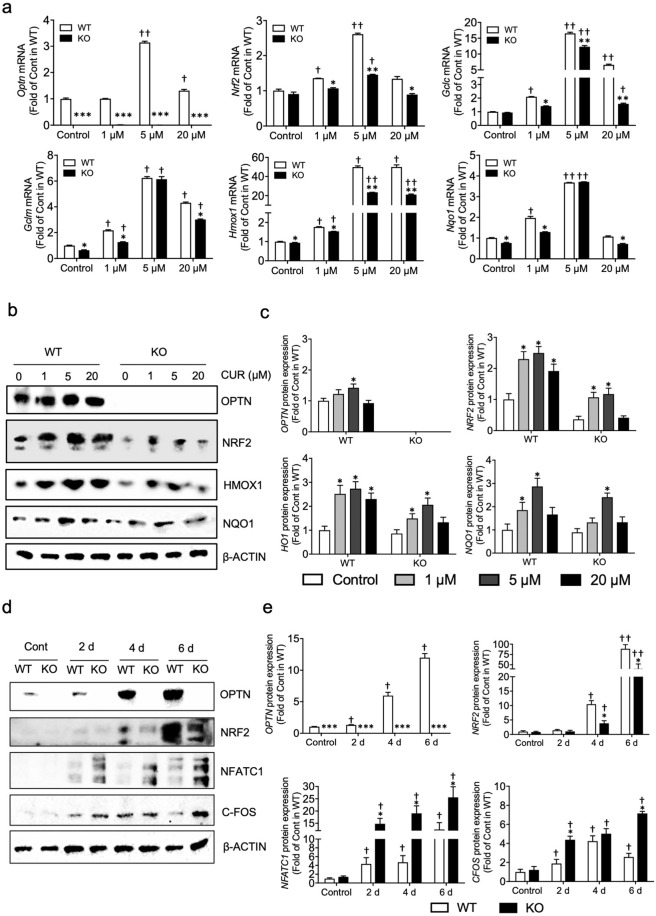
The results described above suggest that the Optn−/− cells are more sensitive to oxidative damage induced by H2O2, indicating impairment of the antioxidative system in these cells. Among the molecules involved in the antioxidant response, NRF2 is considered to be a master transcription factor that can trigger downstream antioxidants to scavenge excess intracellular ROS26,29,45,46. Thus, we next investigated how OPTN deficiency alters antioxidant capability in preosteoclasts. Antioxidant gene expression was first assessed in the Optn−/− and Optn+/+ cells in response to curcumin, a well-known activator of NRF228,47, at concentrations of 1, 5, or 20 µM. As expected, we found no OPTN expression in the Optn−/− cells. Surprisingly, 5 and 20 µM curcumin induced OPTN expression in the Optn+/+ cells (Fig. 4a).
Fig. 4. Deficiency of OPTN attenuates Nrf2 and antioxidant expression.
a Decreased gene expression of Nrf2 and Nrf2 antioxidants in the curcumin-treated Optn−/− vs. Optn+/+ osteoclast precursors. Cells were treated with curcumin (1, 5 and 20 µM) for 6 h. Gene expression of the NRF2-mediated antioxidant genes Hmox1, Gclc, Gclm, and Nqo1 was determined by RT-qPCR with all results were normalized with β-actin. b Decreased protein expression of NRF2 and regulated antioxidants in the curcumin-treated Optn+/+ and Optn−/− preosteoclasts. The cells were treated with curcumin (1, 5 and 20 µM) for 6 h, whole proteins were extracted, and the OPTN, NRF2, HMOX1, and NQO1 levels were measured. c Quantification of WB bands for the protein expression of Nrf2-mediated antioxidants in the curcumin-treated Optn+/+ and Optn−/− preosteoclasts. d Decreased protein levels of NRF2 and key osteoclastogenic markers during osteoclast differentiation in the Optn−/− cells. Primary osteoclast precursors were treated with RANKL (10 ng/ml) and M-CSF (30 ng/mL) for 6 days. At the indicated time points (2, 4, and 6 d), whole protein extracts of the Optn+/+ and Optn−/− cells were extracted and analyzed by Western blotting. e Quantification of WB bands for the protein levels of NRF2 and key osteoclastogenic markers during osteoclast differentiation. CONT, control (growth media only); n = 3 experiments. Data are presented as the mean ± SEM. (a): *p < 0.05, **p < 0.01, ***p < 0.001 compared to Optn+/+ within each dosage group; †p < 0.05, ††p < 0.01 compared to the untreated control of either the Optn+/+ or Optn−/− group; (c): *p < 0.05 compared to the untreated control of either the Optn+/+ or Optn−/− group; e: *p < 0.05, **p < 0.01, ***p < 0.001 compared to Optn+/+ within each time point group; †p < 0.05, ††p < 0.01 compared to the untreated control of either the Optn+/+ or Optn−/− group.
At basal levels, Nrf2 expression trended lower in the Optn−/− vs. Optn+/+ cells, while the mRNA level was significantly decreased in the Optn−/− vs. Optn+/+ cells for the NRF2-mediated antioxidants Gclc, Gclm, Hmox1, and Nqo1. A similar pattern of decreased antioxidant gene expression was also observed in both liver tissues and bone marrow myeloid cells from the Optn−/− animals (Supplementary Fig. S1). Under the treatment conditions used here, Nrf2 was induced by curcumin in the Optn+/+ cells and showed a peak of expression at 5 µM curcumin but was induced to a lesser extent in the Optn−/− cells. With respect to NRF2-mediated antioxidants, expression was slightly increased in both genotypes when exposed to 1 µM curcumin, while the Optn−/− cells exhibited lower antioxidant expression. In the 5 µM curcumin treatment group, all the antioxidants were significantly increased in both the Optn+/+ and Optn−/− cells compared to the controls. However, the Hmox-1 and Gclc levels were significantly lower in the Optn−/− cells than in the Optn+/+ cells, while the Gclm and Nqo1 levels showed a decreasing trend (Fig. 4a). Similarly, in the 20 µM curcumin treatment group, significantly lower antioxidant expression was observed in the Optn−/− cells than in the Optn+/+ cells.
We next assessed the protein expression of NRF2 and its mediated antioxidants. Western blot analysis revealed a similar trend in that the NRF2, HMOX1, and NQO1 levels were lower in the Optn−/− than Optn+/+ cells under basal or/and curcumin-treated conditions (Fig. 4b, c). Although phosphorylation of NRF2 at multiple sites has a limited contribution to modulating the Nrf2-dependent antioxidant response48, we tested this possibility by performing Western blotting for phosphorylated NRF2 (p-NRF2). There was no significant difference in the expression of p-NRF2 at Ser40 between wild-type OPTN−/− preosteoclasts or mature osteoclasts (Supplementary Fig. S2a). These results suggested that OPTN deficiency downregulates NRF2 expression, results in a lack of phosphorylation of NRF2, and attenuates the expression of Nrf2-induced antioxidants. To further evaluate whether NRF2 participates in the process of osteoclastogenesis, we measured the NRF2 protein levels in both the Optn−/− and Optn+/+ cells during the differentiation of precursors into mature osteoclasts (Fig. 4d, e). OPTN protein increased during osteoclastogenesis in the Optn+/+ cells, and while the NRF2 protein levels increased in both cell types, expression in the Optn−/− cells was significantly lower than that in the Optn+/+ cells. Both the gene and protein levels of antioxidants were also decreased (Supplementary Fig. S2a, c). These findings were observed in preosteoclasts during differentiation into mature osteoclasts. The expression of osteoclastogenic markers, including NFATC1 and C-FOS, was elevated in the Optn−/− cells compared to the Optn+/+ cells during osteoclastogenesis, consistent with TRAP staining findings (Fig. 1b). These collective data strongly indicated that OPTN deficiency enhances osteoclastogenesis via a decreased NRF2-mediated antioxidant response, which plays a critical regulatory role in ROS signaling.
Direct interaction between the NRF2 and OPTN proteins
To investigate whether OPTN directly interacts with NRF2, we performed an immunoprecipitation (IP) assay using HEK293T cells. After transient overexpression of FLAG-tagged NRF2 and His-tagged OPTN in cells, we pulled down His-OPTN by anti-FLAG antibodies, suggesting that OPTN and NRF2 can directly interact with each other (Fig. 5a). Both OPTN and NRF2 have been previously identified in the cytoplasm and nucleus36,40,49, and as such, there will be two potential subcellular locations for the OPTN-NRF2 interaction: 1) OPTN may bind to NRF2 in the cytoplasm to facilitate NRF2 translocation or to maintain NRF2 stability against proteasomal degradation50 or 2) OPTN could translocate to the nucleus and bind to NRF2 and other binding partners to form a complex that triggers antioxidant expression.
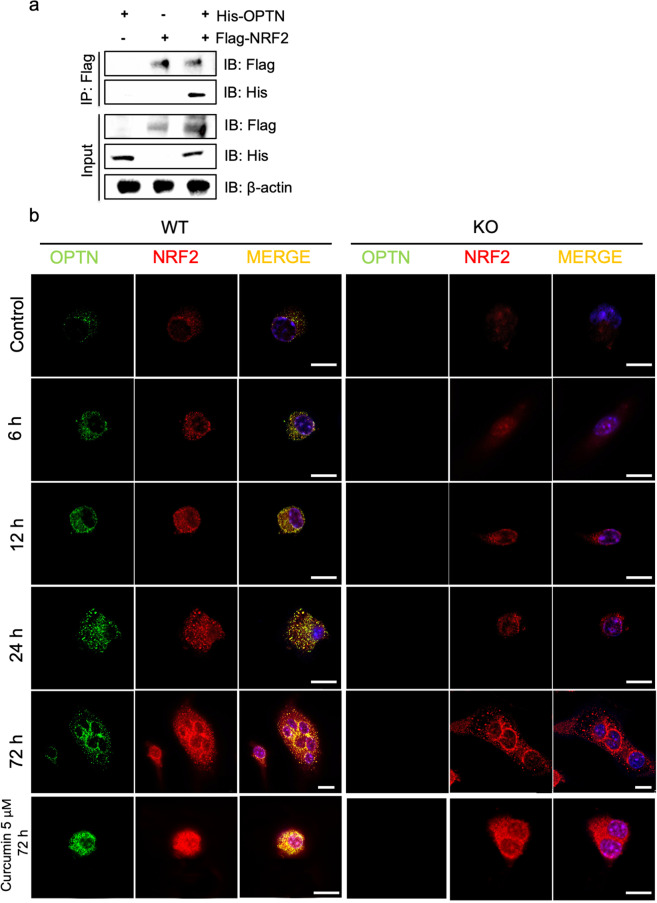
Fig. 5. Direct interaction between the NRF2 and OPTN proteins.
a Immunoblot analysis of whole-cell lysates and CoIP assays using HEK293T cells. Flag immunoprecipitates were isolated from HEK293T cells transfected with His-OPTN or Flag-Nrf2 plasmids for 1 d. The IP bands showed that Flag-tagged NRF2 can pull down His-tagged OPTN, indicating that NRF2 and OPTN can bind with each other in vitro. β-Actin expression served as a loading control. b Intracellular localization of OPTN and NRF2 in preosteoclasts. The Optn−/− and Optn+/+ preosteoclasts were treated with M-CSF (10 ng/mL) and RANKL (30 ng/mL) for 6, 12, 24, and 72 h. RANKL + 5 μM curcumin treatment for 72 h was used as a positive control. After fixation, the cells were processed with immunofluorescence using antibodies against OPTN (green), NRF2 (red) and nuclei (blue). In merged images, colocalization of OPTN and NRF2 was observed mostly in perinuclear granular structures (yellow). Scale bar = 10 μm.
To examine the two possibilities, we visualized the localization of OPTN and NRF2 protein-protein interactions within the cell using immunofluorescence and confocal microscopy. Cells were stimulated with RANKL to initiate osteoclastogenesis, followed by endogenous NRF2 and OPTN immunostaining at various time points (6, 12, 24, and 72 h) after stimulation. The OPTN and NRF2 binding proteins largely localized to the perinuclear region only in the Optn+/+ cells over time. (Fig. 5b). Surprisingly, we also observed a minor degree of colocalization of OPTN and NRF2 in the nucleus, suggesting that a small amount of nuclear OPTN could bind to NRF2, which is consistent with previous reports that OPTN was detected in the nucleus under either stress or overexpression conditions42,49. However, our results indicated that the amount of OPTN- and NRF2-binding proteins in the nucleus was much lower than that in the cytoplasm, indicating that the cytoplasmic interaction between OPTN and NRF2 is the likely dominant binding pattern between OPTN and NRF2 in preosteoclasts during osteoclastogenesis. To our knowledge, this is the first report that OPTN can directly interact with NRF2 predominantly in the perinuclear region in preosteoclasts.
To further elucidate these two possibilities, we performed immunoblotting for NRF2 after separating cytoplasmic and nuclear lysate fractions. We found that NRF2 was decreased in both the cytoplasm and nuclei of the Optn−/− cells. Moreover, the expression of NRF2 in the Optn−/− cells was reduced in nuclear lysates compared to cytoplasmic lysates (Supplementary Fig. S2b). Costaining and quantification of NRF2 and DAPI in osteoclasts also confirmed that nuclear translocation and nuclear expression of NRF2 were significantly lower in the Optn−/− cells during osteoclastogenesis (Supplementary Fig. S2c, d). We then examined NRF2 degradation. We found no difference between the two types of cells in terms of NRF2 degradation following cycloheximide (protein synthesis inhibitor) treatment during osteoclastogenesis (Supplementary Fig. S3c). The Nrf2 Keap1 signaling pathway leads to degradation of NRF2 by the NRF2-KEAP1 interaction, while the noncanonical NRF2-P62 interaction leads to degradation of KEAP1 to allow NRF2 translocation51. We found no difference in KEAP1 expression (Supplementary Fig. S2a) or the NRF2-KEAP1 or NRF2-P62 protein-protein interactions (Supplementary Fig. S3a, b) between the two types of osteoclasts. Collectively, these data showed that OPTN is not involved in NRF2 degradation. Finally, we found no difference in the transcriptional activity of NRF2 between the two types of cells (Supplementary Fig. S3d). In general, our data suggested that OPTN binds to NRF2 in the cytoplasm and may facilitate its nuclear translocation but does not disturb KEAP1-mediated NRF2 degradation and NRF2 transcriptional activity.
NRF2 activators attenuate increased osteoclastogenesis associated with OPTN deficiency
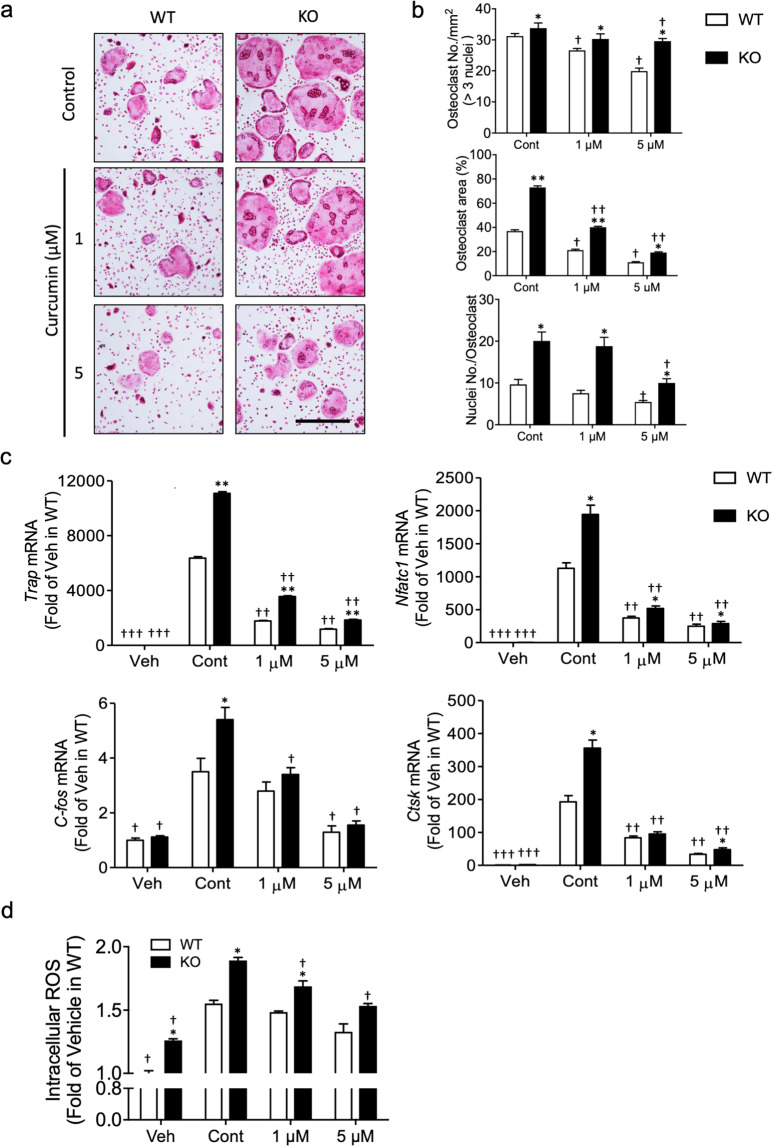
The enhanced antioxidant expression in the Optn−/− cells due to curcumin treatment (Fig. 4a, b) shows the potential for therapeutic applications, suggesting that treatment with NRF2 activators might attenuate elevated osteoclastogenesis induced by OPTN deficiency. To test this possibility, we treated the Optn−/− and Optn+/+ cells with low (1 µM) and high (5 µM) concentrations of curcumin during osteoclastogenesis. Curcumin exhibited potent inhibition of increased osteoclastogenesis in the Optn−/− cells (Fig. 6a), reduced osteoclast number per area, osteoclast size, and number of nuclei per cell in both Optn+/+ and Optn−/− cells, and attenuated elevated osteoclast characteristics in the Optn−/− vs. Optn+/+ cells (Fig. 6b). Curcumin treatment dramatically decreased the expression of critical osteoclastogenic markers, including Trap, Nfatc1, C-fos, and Ctsk, in both the Optn+/+ and Optn−/− cells at the terminal stage of osteoclastogenesis (Fig. 6c). In addition to curcumin, several other NRF2 activators, including quercetin, CDDO-Me, sulforaphane, and tBHQ, showed similar effects on osteoclastogenesis (Supplementary Fig. S4).
Fig. 6. NRF2 activators attenuate increased osteoclastogenesis associated with OPTN deficiency.
a Osteoclastogenesis is enhanced in the Optn−/− cells but is reversed by curcumin treatment of osteoclast precursors. Cells were treated with RANKL/M-CSF together with various concentrations of curcumin (1 and 5 µM) for 5 days. Mature osteoclasts were identified by TRAP staining (red). Scale bar = 500 µm. b Osteoclast number per mm2, the percentage of osteoclast area per image, and nucleus number per osteoclast were quantified based on the images of Fig. 6A. c Osteoclastogenic gene expression in the curcumin-treated preosteoclasts. d Curcumin decreases intracellular ROS levels in the RANKL-treated preosteoclasts. n = 3 experiments. *p < 0.05, **p < 0.01, ***p < 0.001 compared to Optn+/+ within each dosage group; †p < 0.05, ††p < 0.01, †††p < 0.001 compared to the untreated control of either the Optn+/+ or Optn−/− group.
To further elucidate the inhibitory effects of curcumin on osteoclast differentiation, we measured intracellular ROS levels upon curcumin application at the early stage of osteoclastogenesis and found that curcumin significantly attenuated RANKL-stimulated ROS elevation in both the Optn+/+ and Optn−/− cells (Fig. 6d), suggesting that curcumin activated the NRF2-mediated antioxidant response that in turn blocked ROS signaling by scavenging RANKL-induced intracellular ROS. ROS levels in the Optn−/− cells were significantly higher than those in the Optn+/+ cells in all the treatment groups, probably due to the reduced basal antioxidative capability in Optn−/− cells. Taken together, these results suggest that curcumin and other NRF2 activators represent candidates for the treatment of OPTN-associated PDB and demonstrate that NRF2 plays a critical role in OPTN deficiency-induced hyperactive osteoclastogenesis.
Discussion
Optineurin (OPTN) has been recently implicated in the differentiation of osteoclasts; however, the mechanisms identified to date by which OPTN deficiency results in elevated osteoclastogenesis have been limited to the NF-kB and interferon signaling pathways36,52. The different approaches to achieve OPTN deletion in animal models in these studies also contribute to confusion about the relevant underlying mechanisms. Our previous study showed that global deletion of OPTN in mice altered type I interferon signaling but not NF-κB signaling, yielding in vivo features, including osteolytic lesions that result from enhanced osteoclast differentiation and activity36. In this study, we identified an alternative molecular mechanism in which OPTN acts as a binding partner of NRF2, a transcription factor that modulates ROS homeostasis by regulating antioxidant expression. We showed that OPTN modulates the antioxidant response in osteoclasts, demonstrating for the first time that OPTN acts as a negative regulator of osteoclastogenesis by directly interacting with NRF2 (Fig. 7). Importantly, we showed that antioxidant treatment could attenuate OPTN deficiency-associated hyperactive osteoclastogenesis, suggesting to therapeutic possibilities of targeting this pathway.
Fig. 7. Model of hypothesized OPTN regulation of NRF2, ROS, and osteoclastogenesis.
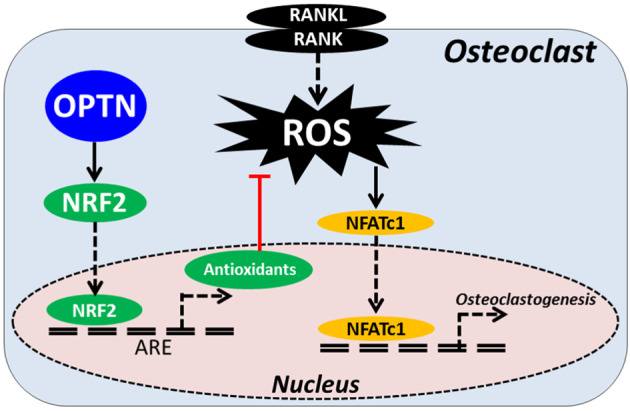
RANKL treatment generates ROS secondary messengers that activate NFATc1 and other genes involved in osteoclastogenesis. ROS homeostasis, including ROS signaling, is maintained by antioxidants, which are in turn transcriptionally regulated by NRF2 and modulated by OPTN.
Our discovery of a direct interaction between OPTN and NRF2 was based on reports that both OPTN and NRF2 contain a basic domain leucine zipper (bZIP) domain, a key feature of transcription factors that are capable of mediating dimerization, transcriptional regulation, and DNA binding53. This finding is consistent with a recent study that showed that OPTN can interact with the bZIP transcription factor neural retina leucine zipper (NRL) in the nucleus of HeLaS3 cells42. Immunoprecipitation demonstrated a direct OPTN-NRF2 interaction, and we also found through immunofluorescence staining of endogenous OPTN and NRF2 that they were colocalized primarily in the perinuclear cytoplasm in preosteoclasts during osteoclastogenesis. At present, the nature of these perinuclear OPTN-NRF2 complexes is unclear, as is their role in regulating downstream NRF2-responsive antioxidant genes. These complexes might be involved in modulating protein transport, degradation, or activity. These possibilities were investigated, and our data suggested that NRF2 nuclear translocation was compromised. Neither NRF2 degradation nor transcriptional activity was affected in the Optn−/− cells. Consistent with our results, Nrf2 global knockout mice have been reported to exhibit augmented intracellular ROS, enhanced RANKL-induced osteoclastogenesis, and increased osteoclastic gene expression due to the impaired antioxidant response28. Future work will be required to elucidate the nature of the OPTN-NRF2 interactions and the mechanism of how this interaction regulates the antioxidant response.
It has been reported that osteoclasts in Nrf2 global knockout mice exhibit augmented intracellular ROS, enhanced RANKL-induced osteoclastogenesis, and increased osteoclastic gene expression due to the impairment of the antioxidant response28. In addition, our previous study identified the dual roles of the NRF2-mediated cellular adaptive response in RANKL-induced osteoclastogenesis, including the protective roles of cells against oxidative damage and the inhibitory role of RANKL-stimulated ROS signaling54. Based on these findings, we further evaluated the treatment effects of multiple compounds that can activate NRF2 in the Optn−/− cells. Notably, all of these compounds showed a strong ability to attenuate the enhanced osteoclastogenesis in the Optn−/− cells, suggesting that these types of NRF2 activators represent novel candidates for the prevention and treatment of diseases involving hyperactivated osteoclasts, such as PDB. Furthermore, these findings support our hypothesis that OPTN regulates osteoclast differentiation by mediating ROS signaling.
The mechanism by which osteoclastogenesis and bone loss increase with aging in vivo remains unclear. The genetic association of PDB with OPTN allowed the development of a genetic PDB mouse model to investigate this mechanism, which we have previously reported36. Although this model exhibited age-dependent bone degeneration consistent with PDB, it remains unclear whether deficiency of the antioxidant response plays a role in vivo. Because it has been reported that uncontrolled ROS will induce progressive cellular damage during the aging process55, we hypothesized that this accumulation of ROS with age and a concomitant decrease in antioxidants may result in elevated osteoclastogenesis, which ultimately causes the PDB-like phenotype in our Optn−/− mouse model. However, given that the pathogenesis of PDB is clearly complex, multifactorial, and time-dependent, the underlying mechanisms by which OPTN plays a role in the process require further investigation.
While physiological levels of ROS can serve as secondary messengers in multiple signaling cascades54,56,57, excessive ROS lead to oxidative stress and, if not contained, will eventually damage lipids, proteins, and DNA, resulting in oxidative damage. Our data (Fig. 3) showed that the Optn−/− cells are more susceptible to the cytotoxicity induced by high levels of H2O2 than the control cells, which is probably due to the attenuated antioxidative capacities of cells. This decrease in antioxidants and a concomitant accumulation of ROS with age could be a potential explanation for this age-dependent bone loss. Moreover, uncontrolled ROS accumulation with aging and the resulting oxidative damage have been implicated in various disorders, such as cancer, neurodegeneration, and other age-dependent diseases58. Our discovery that OPTN regulates ROS through NRF2-dependent antioxidants may have substantial translational and clinical significance beyond disorders of bone degeneration. OPTN has also been genetically associated with neurodegeneration in glaucoma and ALS30–32, suggesting that our findings may contribute to novel therapies for neurodegenerative diseases.
Supplementary information
Acknowledgements
This work was supported, in part, by NIH/NIDCR R01DE022816 and the Ohio State University College of Dentistry.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Peng Xue, Xiangxiang Hu
Supplementary information
The online version contains supplementary material available at 10.1038/s12276-021-00596-w.
References
- 1.Seeman E. Bone modeling and remodeling. Crit. Rev. Eukaryot. Gene Expr. 2009;19:219–233. doi: 10.1615/critreveukargeneexpr.v19.i3.40. [DOI] [PubMed] [Google Scholar]
- 2.Kylmaoja E, Nakamura M, Tuukkanen J. Osteoclasts and remodeling based bone formation. Curr. Stem Cell Res Ther. 2016;11:626–633. doi: 10.2174/1574888x10666151019115724. [DOI] [PubMed] [Google Scholar]
- 3.Teitelbaum SL. Osteoclasts: what do they do and how do they do it? Am. J. Pathol. 2007;170:427–435. doi: 10.2353/ajpath.2007.060834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner JM, et al. Inflammatory processes and elevated osteoclast activity chaperon atrophic non-union establishment in a murine model. J. Transl. Med. 2019;17:416. doi: 10.1186/s12967-019-02171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silva IAL, et al. Effect of genetic variants of OPTN in the pathophysiology of Paget’s disease of bone. Biochim Biophys. Acta. 2018;1864:143–151. doi: 10.1016/j.bbadis.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Alonso N, Calero-Paniagua I, Del Pino-Montes J. Clinical and genetic advances in paget’s disease of bone: a review. Clin. Rev. Bone Min. Metab. 2017;15:37–48. doi: 10.1007/s12018-016-9226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nebot Valenzuela E, Pietschmann P. Epidemiology and pathology of Paget’s disease of bone - a review. Wien. Med Wochenschr. 2017;167:2–8. doi: 10.1007/s10354-016-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vallet M, Ralston SH. Biology and treatment of Paget’s disease of bone. J. Cell Biochem. 2016;117:289–299. doi: 10.1002/jcb.25291. [DOI] [PubMed] [Google Scholar]
- 9.Brown C. Osteoporosis: Staying strong. Nature. 2017;550:S15–S17. doi: 10.1038/550S15a. [DOI] [PubMed] [Google Scholar]
- 10.Ralston SH, Langston AL, Reid IR. Pathogenesis and management of Paget’s disease of bone. Lancet. 2008;372:155–163. doi: 10.1016/S0140-6736(08)61035-1. [DOI] [PubMed] [Google Scholar]
- 11.Park JH, Lee NK, Lee SY. Current understanding of RANK signaling in osteoclast differentiation and maturation. Mol. Cells. 2017;40:706–713. doi: 10.14348/molcells.2017.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li DZ, Zhang QX, Dong XX, Li HD, Ma X. Treatment with hydrogen molecules prevents RANKL-induced osteoclast differentiation associated with inhibition of ROS formation and inactivation of MAPK, AKT and NF-kappa B pathways in murine RAW264.7 cells. J. Bone Min. Metab. 2014;32:494–504. doi: 10.1007/s00774-013-0530-1. [DOI] [PubMed] [Google Scholar]
- 13.Kim MS, et al. RANKL-mediated reactive oxygen species pathway that induces long lasting Ca2+ oscillations essential for osteoclastogenesis. J. Biol. Chem. 2010;285:6913–6921. doi: 10.1074/jbc.M109.051557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee NK, et al. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood. 2005;106:852–859. doi: 10.1182/blood-2004-09-3662. [DOI] [PubMed] [Google Scholar]
- 15.Kim HS, et al. DJ-1 controls bone homeostasis through the regulation of osteoclast differentiation. Nat. Commun. 2017;8:1519. doi: 10.1038/s41467-017-01527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 17.Giacco F, Brownlee M. Oxidative stress and diabetic complications. Circ. Res. 2010;107:1058–1070. doi: 10.1161/CIRCRESAHA.110.223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Tan J, Miao Y, Lei P, Zhang Q. ROS and autophagy: interactions and molecular regulatory mechanisms. Cell Mol. Neurobiol. 2015;35:615–621. doi: 10.1007/s10571-015-0166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaarniranta, K., Pawlowska, E., Szczepanska, J., Jablkowska, A. & Blasiak, J. Role of mitochondrial DNA damage in ROS-mediated pathogenesis of age-related macular degeneration (AMD). Int. J. Mol. Sci.10.3390/ijms20102374 (2019). [DOI] [PMC free article] [PubMed]
- 20.Li PL, Gulbins E. Bioactive lipids and redox signaling: molecular mechanism and disease pathogenesis. Antioxid. Redox Signal. 2018;28:911–915. doi: 10.1089/ars.2017.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi Y, et al. Dimethyl fumarate inhibits osteoclasts via attenuation of reactive oxygen species signalling by augmented antioxidation. J. Cell Mol. Med. 2018;22:1138–1147. doi: 10.1111/jcmm.13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thummuri D, Naidu VGM, Chaudhari P. Carnosic acid attenuates RANKL-induced oxidative stress and osteoclastogenesis via induction of Nrf2 and suppression of NF-kappaB and MAPK signalling. J. Mol. Med (Berl.) 2017;95:1065–1076. doi: 10.1007/s00109-017-1553-1. [DOI] [PubMed] [Google Scholar]
- 23.Kim H, Lee YD, Kim HJ, Lee ZH, Kim HH. SOD2 and Sirt3 control osteoclastogenesis by regulating mitochondrial ROS. J. Bone Min. Res. 2017;32:397–406. doi: 10.1002/jbmr.2974. [DOI] [PubMed] [Google Scholar]
- 24.Chen, Y. et al. Alliin attenuated RANKL-induced osteoclastogenesis by scavenging reactive oxygen species through inhibiting Nox1. Int. J. Mol. Sci.10.3390/ijms17091516 (2016). [DOI] [PMC free article] [PubMed]
- 25.Dou C, et al. Cordycepin prevents bone loss through inhibiting osteoclastogenesis by scavenging ROS generation. Nutrients. 2016;8:231. doi: 10.3390/nu8040231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanzaki H, Shinohara F, Itohiya-Kasuya K, Ishikawa M, Nakamura Y. Nrf2 activation attenuates both orthodontic tooth movement and relapse. J. Dent. Res. 2015;94:787–794. doi: 10.1177/0022034515577814. [DOI] [PubMed] [Google Scholar]
- 27.Kanzaki H, Shinohara F, Kajiya M, Kodama T. The Keap1/Nrf2 protein axis plays a role in osteoclast differentiation by regulating intracellular reactive oxygen species signaling. J. Biol. Chem. 2013;288:23009–23020. doi: 10.1074/jbc.M113.478545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hyeon S, Lee H, Yang Y, Jeong W. Nrf2 deficiency induces oxidative stress and promotes RANKL-induced osteoclast differentiation. Free Radic. Biol. Med. 2013;65:789–799. doi: 10.1016/j.freeradbiomed.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Xue, P. et al. CDDO-Me, Sulforaphane and tBHQ attenuate the RANKL-induced osteoclast differentiation via activating the NRF2-mediated antioxidant response. Biochem. Biophys. Res. Commun.10.1016/j.bbrc.2019.02.095 (2019). [DOI] [PMC free article] [PubMed]
- 30.Minegishi Y, Nakayama M, Iejima D, Kawase K, Iwata T. Significance of optineurin mutations in glaucoma and other diseases. Prog. Retin Eye Res. 2016;55:149–181. doi: 10.1016/j.preteyeres.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Morimoto S, et al. Optineurin pathology in the spinal cord of amyotrophic lateral sclerosis/parkinsonism-dementia complex patients in Kii Peninsula, Japan. Brain Pathol. 2018;28:422–426. doi: 10.1111/bpa.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Blitterswijk M, et al. Novel optineurin mutations in sporadic amyotrophic lateral sclerosis patients. Neurobiol. Aging. 2012;33:1016 e1011–1016 e1017. doi: 10.1016/j.neurobiolaging.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 33.Albagha OM, et al. Genome-wide association study identifies variants at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget’s disease of bone. Nat. Genet. 2010;42:520–524. doi: 10.1038/ng.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung PY, et al. The majority of the genetic risk for Paget’s disease of bone is explained by genetic variants close to the CSF1, OPTN, TM7SF4, and TNFRSF11A genes. Hum. Genet. 2010;128:615–626. doi: 10.1007/s00439-010-0888-2. [DOI] [PubMed] [Google Scholar]
- 35.Silva IAL, et al. Effect of genetic variants of OPTN in the pathophysiology of Paget’s disease of bone. Biochim Biophys. Acta Mol. Basis Dis. 2018;1864:143–151. doi: 10.1016/j.bbadis.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Wong SW, et al. Global deletion of Optineurin results in altered type I IFN signaling and abnormal bone remodeling in a model of Paget’s disease. Cell Death Differ. 2020;27:71–84. doi: 10.1038/s41418-019-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, Ohtsubo M, Hotta Y, Minoshima S. Oligomerization of optineurin and its oxidative stress- or E50K mutation-driven covalent cross-linking: possible relationship with glaucoma pathology. PLoS One. 2014;9:e101206. doi: 10.1371/journal.pone.0101206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu M, Li A, Chen J, Zhang S, Wu J. Effects of optineurin mutants on SH-SY5Y cell survival. Mol. Cell Neurosci. 2016;74:18–24. doi: 10.1016/j.mcn.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Kato K, et al. Relationship between radiosensitivity and Nrf2 target gene expression in human hematopoietic stem cells. Radiat. Res. 2010;174:177–184. doi: 10.1667/RR2146.1. [DOI] [PubMed] [Google Scholar]
- 40.Itoh K, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 41.Plafker KS, et al. The ubiquitin-conjugating enzyme UbcM2 can regulate the stability and activity of the antioxidant transcription factor Nrf2. J. Biol. Chem. 2010;285:23064–23074. doi: 10.1074/jbc.M110.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C, et al. Interaction between optineurin and the bZIP transcription factor NRL. Cell Biol. Int. 2014;38:16–25. doi: 10.1002/cbin.10174. [DOI] [PubMed] [Google Scholar]
- 43.Reinke AW, Baek J, Ashenberg O, Keating AE. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science. 2013;340:730–734. doi: 10.1126/science.1233465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SY, Lee KS, Yi SH, Kook SH, Lee JC. Acteoside suppresses RANKL-mediated osteoclastogenesis by inhibiting c-Fos induction and NF-kappaB pathway and attenuating ROS production. PLoS One. 2013;8:e80873. doi: 10.1371/journal.pone.0080873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahmed SM, Luo L, Namani A, Wang XJ, Tang X. Nrf2 signaling pathway: pivotal roles in inflammation. Biochim Biophys. Acta Mol. Basis Dis. 2017;1863:585–597. doi: 10.1016/j.bbadis.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Kanzaki H, et al. Nuclear Nrf2 induction by protein transduction attenuates osteoclastogenesis. Free Radic. Biol. Med. 2014;77:239–248. doi: 10.1016/j.freeradbiomed.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Zhao R, et al. Curcumin protects human keratinocytes against inorganic arsenite-induced acute cytotoxicity through an NRF2-dependent mechanism. Oxid. Med. Cell. Longev. 2013;2013:412576. doi: 10.1155/2013/412576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Z, Huang Z, Zhang DD. Phosphorylation of Nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the Nrf2-dependent antioxidant response. PLoS One. 2009;4:e6588. doi: 10.1371/journal.pone.0006588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Marco N, Buono M, Troise F, Diez-Roux G. Optineurin increases cell survival and translocates to the nucleus in a Rab8-dependent manner upon an apoptotic stimulus. J. Biol. Chem. 2006;281:16147–16156. doi: 10.1074/jbc.M601467200. [DOI] [PubMed] [Google Scholar]
- 50.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah SZA, et al. p62-Keap1-NRF2-ARE pathway: a contentious player for selective targeting of autophagy, oxidative stress and mitochondrial dysfunction in prion diseases. Front Mol. Neurosci. 2018;11:310. doi: 10.3389/fnmol.2018.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Obaid R, et al. Optineurin negatively regulates osteoclast differentiation by modulating NF-kappaB and interferon signaling: implications for Paget’s disease. Cell Rep. 2015;13:1096–1102. doi: 10.1016/j.celrep.2015.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tonelli C, Chio IIC, Tuveson DA. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018;29:1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xue P, et al. CDDO-Me, Sulforaphane and tBHQ attenuate the RANKL-induced osteoclast differentiation via activating the NRF2-mediated antioxidant response. Biochem Biophys. Res Commun. 2019;511:637–643. doi: 10.1016/j.bbrc.2019.02.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stefanatos R, Sanz A. The role of mitochondrial ROS in the aging brain. FEBS Lett. 2018;592:743–758. doi: 10.1002/1873-3468.12902. [DOI] [PubMed] [Google Scholar]
- 56.Xue P, et al. Prolonged inorganic arsenite exposure suppresses insulin-stimulated AKT S473 phosphorylation and glucose uptake in 3T3-L1 adipocytes: involvement of the adaptive antioxidant response. Biochem Biophys. Res Commun. 2011;407:360–365. doi: 10.1016/j.bbrc.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng H, et al. CNC-bZIP protein Nrf1-dependent regulation of glucose-stimulated insulin secretion. Antioxid. Redox Signal. 2015;22:819–831. doi: 10.1089/ars.2014.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moreira PI, et al. Oxidative stress and neurodegeneration. Ann. NY Acad. Sci. 2005;1043:545–552. doi: 10.1196/annals.1333.062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.