Figure EV5. sgRNA targeting four isoforms of SMAD3 and BRAFi resistance.
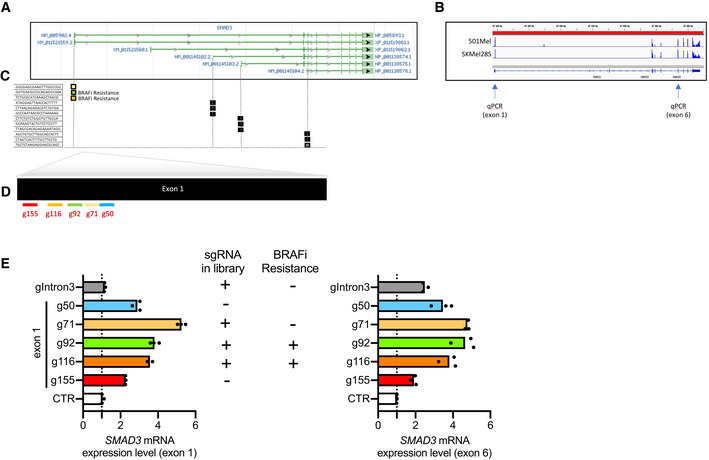
- SMAD3 mRNA isoforms according to NCBI. The longest isoform NM_005902.4 encodes SMAD3 protein (NP_005893.1). Three shorter isoforms of SMAD3 mRNA encode shorter SMAD3 proteins.
- Snapshot of SMAD3 mRNA expression in 501Mel and SKMel28S cells using IGV tool. Note that exon 1 is detectable in these melanoma cell lines. Arrows indicate the binding sites of primers for RT–qPCR (exon 1 or 6 for NM_005902.4).
- Ability of six sgRNAs to transactivate SMAD3 expression have been compared. Among the five sgRNAs targeting the first exon, three are found in the library (g71, g92, and g116). We designed g50 and g155 to evaluate the impact of sgRNA position on exon 1. sgRNA gIntron3 has been designed as a control (unable to promote the longest SMAD3 isoform but efficient to transactivate the smallest isoform). These two isoforms contain the exon 6 used for RT–qPCR.
- SMAD3 quantification by RT–qPCR (exon 1 or 6) in HEK293 cells transiently transfected with the three plasmids required for the CRISPR‐SAM as previously described (Konermann et al, 2015). CTR for empty plasmid (no sgRNA cloned into lenti sgRNA(MS2)_zeo backbone). RT–qPCR data are representative of two independent experiments. Source data are available in Fig EV5 source data. Each histogram represents the mean of three technical replicates from one representative experiment. No statistical analyses were thus performed. Dotted line highlights the value of 1 (corresponding to the effect obtained by the control sgRNA).
Source data are available online for this figure.
