Fig. 2.
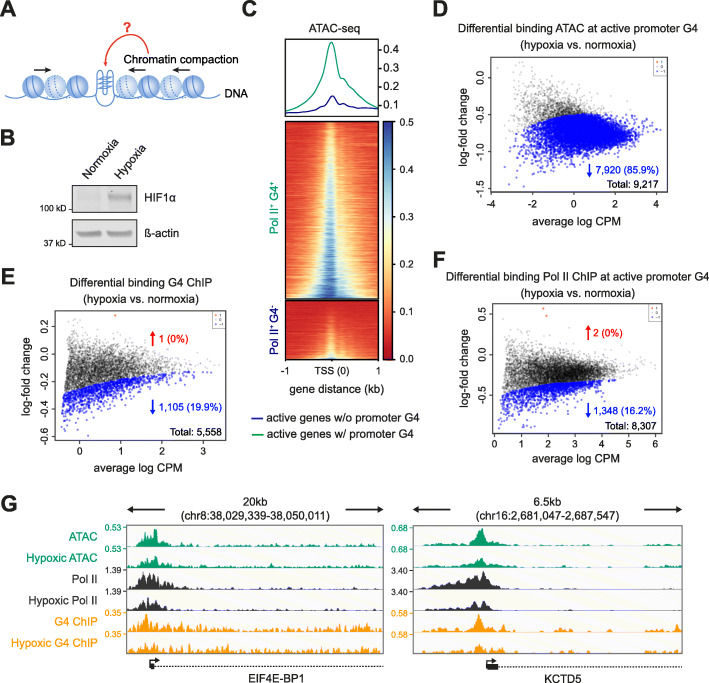
Chromatin compaction diminishes promoter G4 folding. a Graphical representation of experimental design examining G4s, transcription and chromatin compaction induced by hypoxia. b Western blotting for HIF1α from K562 cells cultured under normoxic or hypoxic conditions. ß-actin provides the loading control. c Chromatin accessibility at promoters under normoxic conditions. Top panel, metagene plot for the median of the normalised ATAC signal (3 biological replicates) centred at the TSS under normoxic conditions. Green, promoters of active genes with a G4 (Pol II+ G4+); blue, active genes without a promoter G4 (Pol II+ G4−). Bottom panel, data plotted for individual loci and represented by heatmaps. d–f MA plots showing fold change in ATAC-seq signal (d), G4 ChIP-seq signal (e) and Pol II ChIP-seq signal (f) following hypoxia at active promoters with a G4 (Pol II+ G4+). Blue and red, sites with significantly reduced or increase signal respectively (p < 0.05). CPM read count per million. g Genomic view of EIF4E-BP1 and KCTD5 exemplify genes that significantly change in ATAC-seq (green), Pol II ChIP-seq (black) and G4 ChIP-seq peaks (yellow). Tracks compare peaks between hypoxia and normoxia. Genomic coordinates indicate track range and the signal is quantified as counts per million (CPM). In all panels normoxia refers to cells cultured in 21% O2 and hypoxia refers to cells exposed to 1% O2 for 1 h