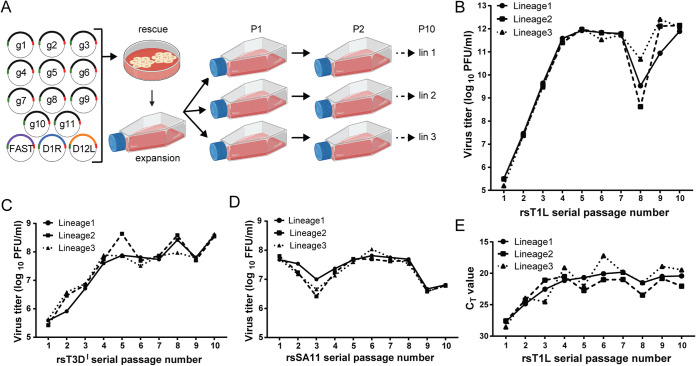
FIG 1.
Serial passage workflow and virus titers. (A) Workflow for rotavirus serial passaging. Baby hamster kidney cells expressing T7 RNA polymerase were transfected with plasmids encoding the 11 rotavirus positive-sense viral RNAs (+RNAs) (g1 to g11) and helper plasmids encoding capping enzymes (D1R and D12L) and a cell-cell fusion protein (FAST) and then cocultured with MA104 cells to promote recombinant virus rescue. rsSA11 was amplified by a single passage in MA104 cells, and the virus stock titer was determined. To generate P1 stocks, MA104 monolayers in three flasks were adsorbed at an MOI of 0.25 PFU/cell, washed, and incubated with fresh medium for 48 h prior to lysis by multiple rounds of freezing and thawing. Subsequent passages (P2 to P10) were generated by adsorption of MA104 monolayers with 3 ml of cleared lysate from the previous passage, with three lineages each passaged in an independent series. A similar workflow was used for rescue and passaging of reovirus, employing standard reverse genetics approaches (23, 71) and with passages conducted in suspension rather than monolayer culture. Created with BioRender.com. (B and C) Graphs showing titers for three lineages of rsT1L (B) or rsT3DI (C) reoviruses across 10 serial passages, quantified by plaque assay. (D) Graphs showing titers for three lineages of rsSA11 rotavirus across all 10 serial passages, quantified by fluorescent focus assay. (E) Graph showing CT values from S4 RT-qPCR analysis of RNA extracted from three lineages of rsT1L across 10 passages.