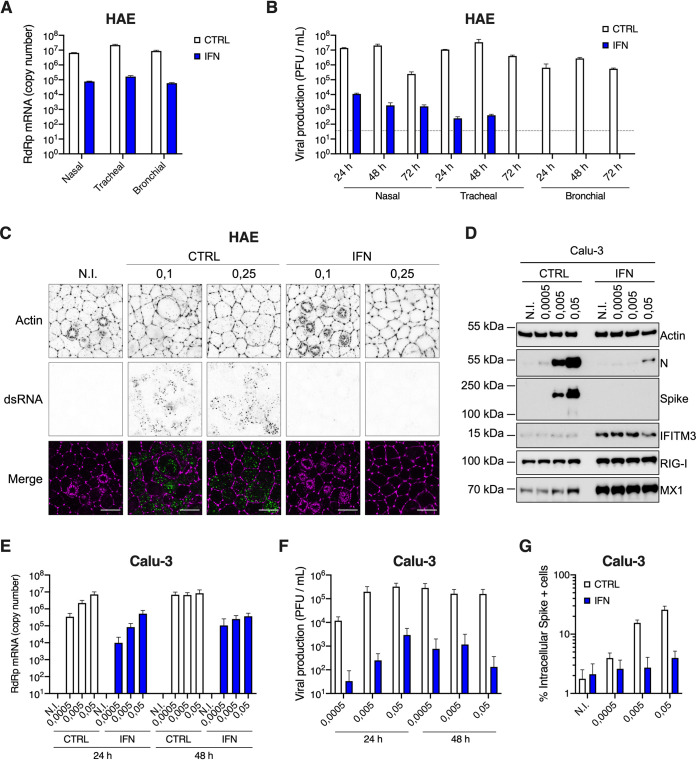
FIG 7.
Inhibition of SARS-CoV-2 replication by type I IFN pretreatment in primary HAE cells and immortalized Calu-3 cells. (A) Human HAE cells (nasal, tracheal, or bronchial, as indicated) were pretreated or not with type I IFN for 20 h, and N.I. or incubated with SARS-CoV-2 on the apical side at MOI 0.1 for 1 to 2 h. Cells were harvested at 72 h postinfection, then lysed for RNA extraction and RdRp RT-qPCR analysis. (B) Plaque assays were performed on washes of the apical side of the HAE cells from (A) at 24 h, 48 h, and 72 h to determine the number of PFU per ml of supernatant (gray dotted line = detection threshold). (C) Human HAE cells were pretreated or not with IFN for 20 h, and N.I. or incubated with SARS-CoV-2 on the apical side at MOI 0.1 and 0.25 for 2 h. After 48 h, cells were stained for actin with phalloidin (magenta) and an anti-double-stranded RNA antibody (green). Representative images, acquired with an LSM880 Airyscan microscope, are shown; scale bar 10 μm. D. Calu-3 cells were pretreated or not with IFN for 16 to 20 h, the cells were N.I. or incubated with SARS-CoV-2 at the indicated MOIs, and lysed 24 h postinfection for immunoblot analysis of SARS-CoV-2 nucleoprotein (N) and spike, and IFITM3, RIG-I, MX1, and actin expression levels. A representative immunoblot is shown. (E) Human Calu-3 cells were pretreated or not with IFN, and infected as in (D). Cells were harvested and lysed for RNA extraction and RdRp RT-qPCR analysis. (F) Production of infectious viruses in supernatants from (E) was determined by plaque assays. (G) Calu-3 cells were pretreated or not with IFN and infected as in (D), and cells were stained with an anti-spike antibody. The percentage of spike positive (+) cells was scored by flow cytometry. The means of three (A and B) or four (E to G) independent experiments are shown, with error bars representing the SD from the mean.