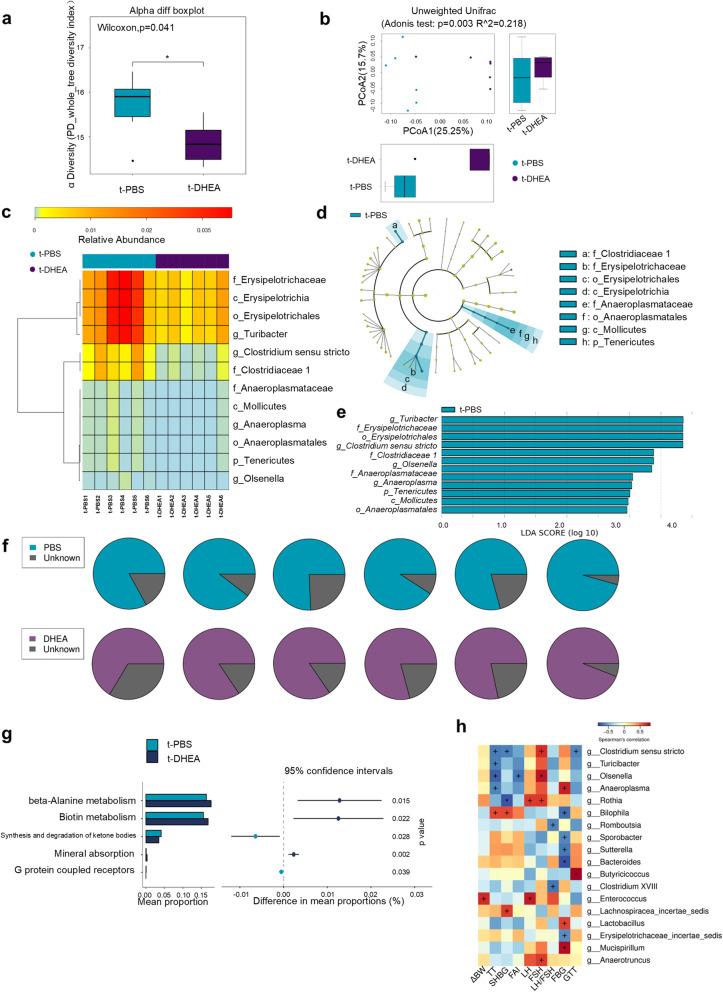
Fig. 3.
Gut microbiota profiling of FMT recipient rats. a Phylogenic diversity whole tree index indicating alpha diversity in the 2 groups. b PCoA analysis with unweighted UniFrac distance. Numbers on the axes represent percentages of variation interpreted by that coordinate. c Wilcoxon test identified differentially abundant taxa in gut microbiota after being transferred from PCOS-like rats and controls. Cladogram (d) and bar plot (e) of LEfSe analysis showed taxa contribution to group separation. Taxa with LDA score > 2 were displayed. f Pie chart of SourceTracker analysis revealing percentages of gut microbiota originated from their corresponding donors. g Differentially enriched modules visualised by STAMP. h Heat map of the Spearman’s rho correlation test presenting the association between genus-level microbes and clinical parameters. Red squares represent positive correlations between relative abundances of microbes and clinical parameters; blue squares represent negative correlations. +, p < 0.10; *, p < 0.05. n = 6 rats per group