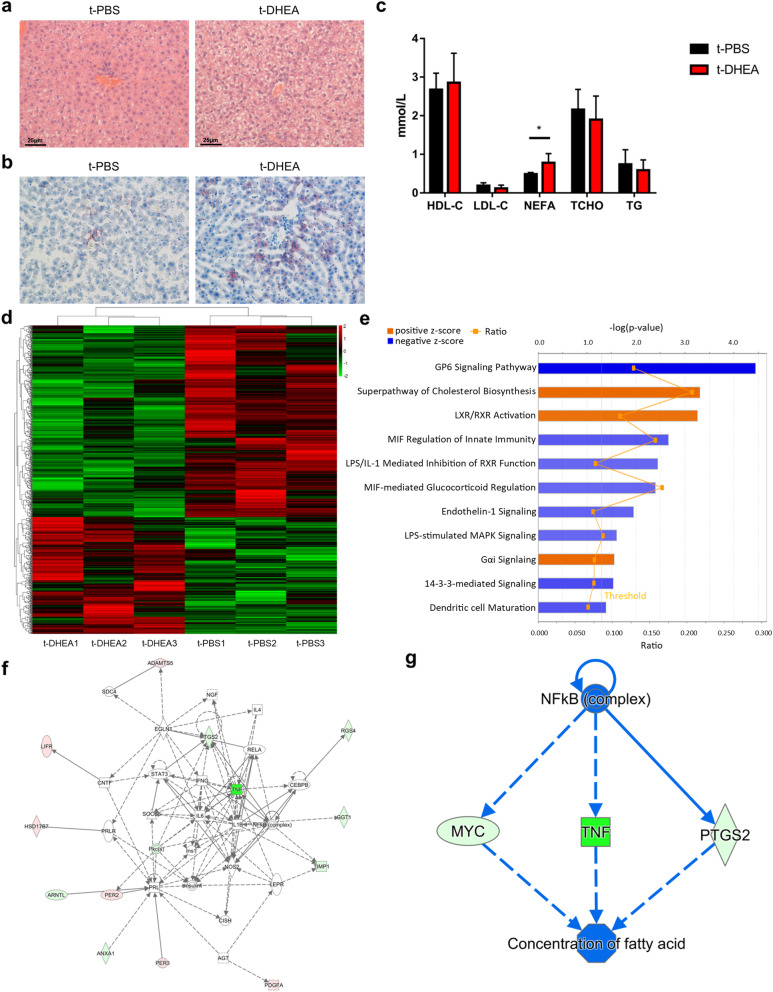
Fig. 5.
FMT of DHEA-shaped gut microbiota regulated liver metabolism. Representative photomicrographs revealing hepatic lipid content by H&E staining (a) and ORO staining (b). Scale bar = 25 μm. c Serum lipid levels including HDL-C, LDL-C, NEFA, TCHO, and TG in t-PBS group and t-DHEA group (n = 6 each group, * p < 0.05). Data are given as mean ± SEM. d Heat map of 725 DEGs in livers from t-PBS and t-DHEA groups (n = 3 each group). These data were presented using FPKM values. e Bar plot summarises the significantly influenced canonical pathways regarding metabolism and immunity in rat livers (n = 3 each group). Pathways with Z-score more than 2 (orange) or less than − 2 (blue) were displayed and were ranked by − log(p value). The straight orange line indicates the significance level of p = 0.05. The orange fold line indicates the ratio of enrichment in the pathway. f Network presenting interactions between DEGs involved in carbohydrate and lipid metabolism (n = 3 each group). Genes are exhibited as red nodes (upregulated) and green nodes (downregulated). Relationships between 2 nodes are indicated as lines (direct interaction) or dotted lines (indirect interaction). g Network displaying regulatory effects from upstream regulators to downstream bio-functions linked by DEGs (n = 3 each group). DEGs, differentially expressed genes; FPKM, Fragments Per Kilobase of exon model per million mapped reads; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; NEFA, non-esterified fatty acid; TCHO, total cholesterol; TG, triglycerides