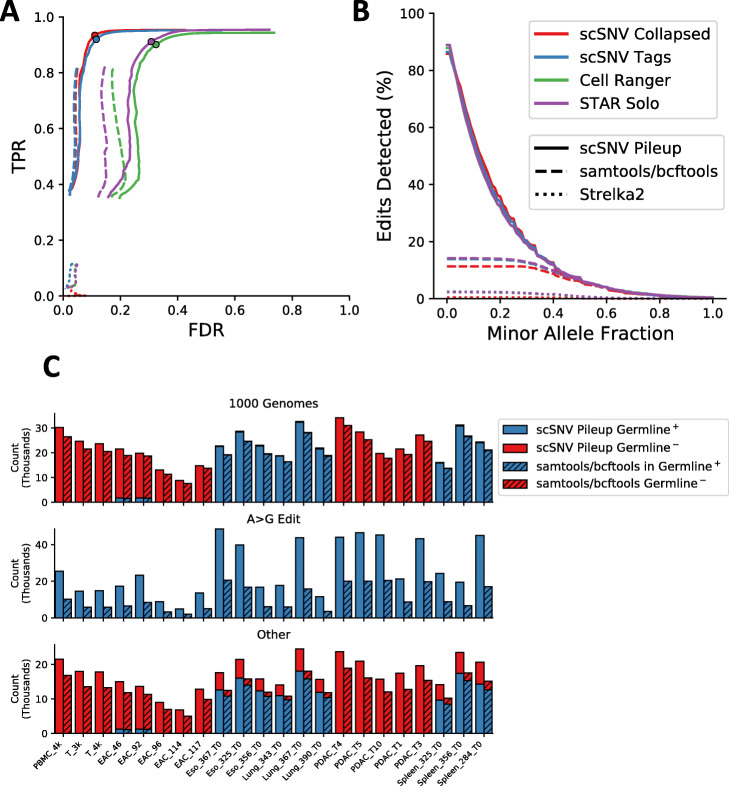
Fig. 3.
Comparison of SNV calling performance using four alignment methods and three SNV calling methods. a The median TPR and FDR rates stratified by increasing minor allele fraction cutoffs estimated using matched DNA sequencing. The color indicates the alignment method and line style indicates the SNV calling method as per the legend. The dots on the lines using scSNV Pileup indicate the position of our recommended minor allele fraction of 0.25. b The number of A>G edits detected with increasing minor allele fraction cutoffs using the same methods as a. c The number of SNV calls (in thousands) for SNVs overlapping 1000 Genomes common SNVs (first row), predicted to be A>G edits (second row), or SNVs without an overlapping annotation (third row) for SNVs identified using scSNV Collapse with scSNV Pileup (solid) or samtools/bcftools (dashed) across twenty-two samples. For samples with matched DNA sequencing data, the number of SNVs found within the germline calls for each method is indicated in blue, while those not found in the germline calls are indicated in red