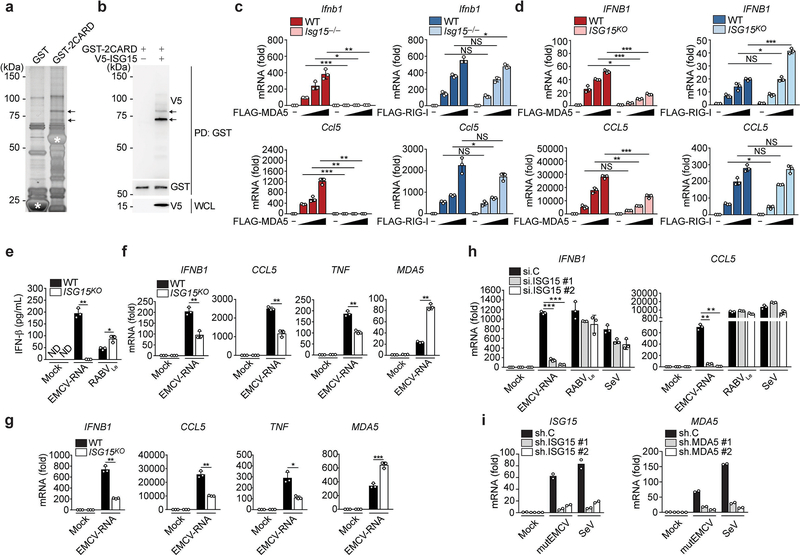
Extended Data Fig. 1. ISG15 is required for MDA5, but not RIG-I, mediated signal transduction.
(a) Silver-stained affinity-purified GST and GST-MDA5–2CARD from transiently transfected HEK293T cells. Asterisks denote the GST and GST-MDA5–2CARD (aa 1–295) proteins. Arrows indicate the bands that identified ISG15 by MS analysis. (b) GST-MDA5–2CARD ISGylation in transiently transfected HEK293T cells with or without co-expressed V5-ISG15, determined by GST pulldown (PD) and immunoblot (IB) with anti-V5 and anti-GST. Whole cell lysates (WCL) were probed by IB with anti-V5. (c, d) qRT-PCR analysis of IFNB1 and CCL5 transcripts in WT and Isg15−/− MEFs (c) or WT and ISG15 KO HeLa cells (d) that were transfected with empty vector or increasing amounts of FLAG-MDA5 or FLAG-RIG-I for 40 h. (e) ELISA of IFN-β in the supernatants of WT or ISG15 KO HeLa cells that were mock-transfected or transfected with EMCV-RNA (0.4 μg/mL) or RABVLe (1 pmol/mL) for 24 h. (f) qRT-PCR analysis of IFNB1, CCL5, TNF, and MDA5 mRNA in WT and ISG15 KO HeLa cells that were mock-transfected or transfected with EMCV-RNA (0.4 μg/mL) for 24 h. (g) qRT-PCR analysis of IFNB1, CCL5, TNF, and MDA5 mRNA in WT and ISG15 KO HAP-1 cells that were stimulated as in (f). (h) qPCR analysis of IFNB1 and CCL5 mRNA in NHLFs that were transfected with the indicated siRNAs for 30 h and then mock-stimulated or transfected with EMCV-RNA (0.4 μg/mL) or RABVLe (1 pmol/mL), or infected with SeV (10 HAU/mL) for 16 h. (i) qRT-PCR analysis of ISG15 and MDA5 mRNA in PBMCs that were transduced for 40 h with the indicated shRNA lentiviral particles and then infected with mutEMCV (MOI 10) or SeV (200 HAU/mL) for 8 h Data are representative of at least two independent experiments (mean ± s.d. of n = 3 biological replicates in c, d, e, f, g, and h; mean of n = 2 biological replicates in i). *p < 0.05, **p < 0.01, ***p < 0.001 (two-tailed unpaired t-test). NS, not significant; ND, not detected.