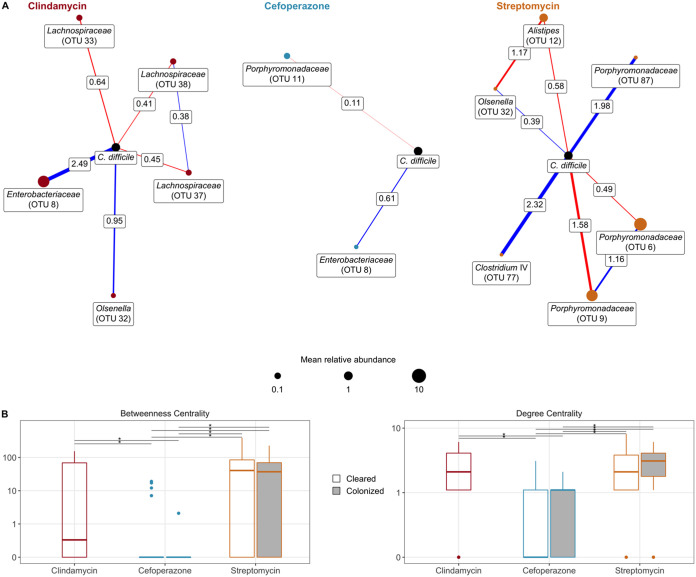
FIG 6.
Conditional independence networks reveal treatment-specific relationships between the community and C. difficile during colonization clearance. (A) Sparse inverse covariance estimation for ecological association inference (SPIEC-EASI) networks showing conditionally independent first-order relationships between C. difficile and the community as C. difficile was cleared from the gut environment. Nodes are sized by the median relative abundance of the OTU. A red edge indicates a negative interaction, and blue indicates a positive interaction, while edge thickness indicates the interaction’s strength. (B) Network centrality measured with betweenness, i.e., how many paths between two OTUs pass through an individual, and degree, i.e., how many connections an OTU has. *, P < 0.05, calculated by Wilcoxon rank sum test with Benjamini-Hochberg correction.