Figure 2.
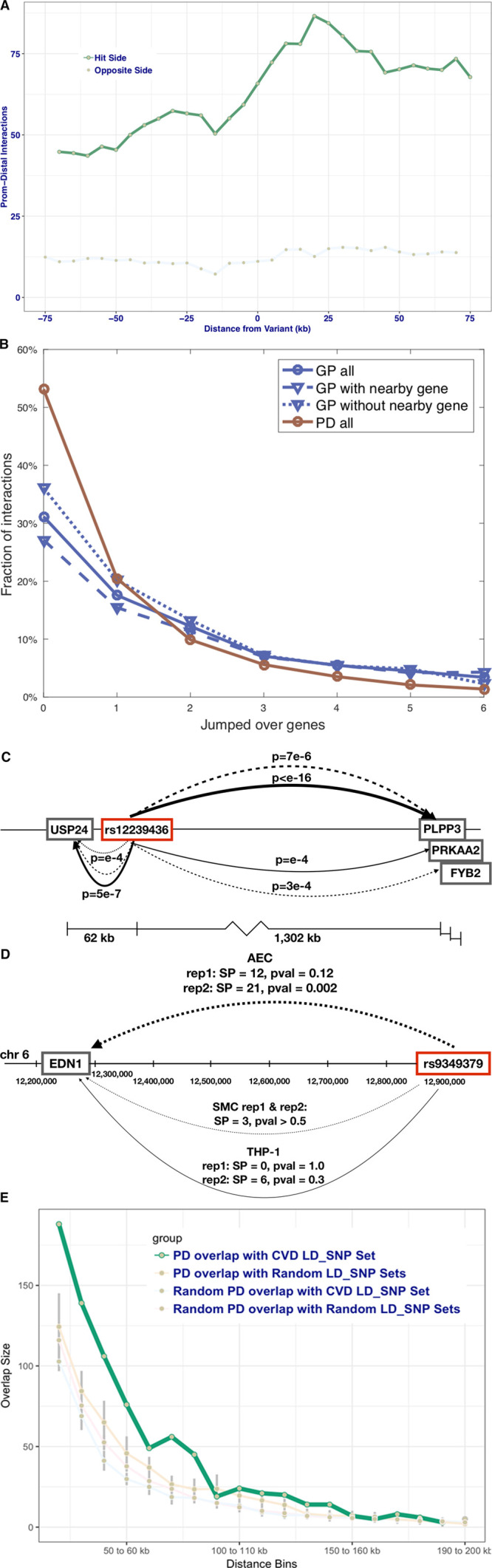
HiCap can inform on regulatory potential of variants in LD with risk variants. A, The aortic endothelial cell (AEC) Prom-Dist (PD) set was searched for interactions between the promoter and distal elements close to a variant interacting with the same promoter (green curve). The comparison was made relative to a site at the same distance from the promoter it but located at the other side of it (light blue curve). The latter is nearly a horizontal line as expected, whereas the blue curve strongly deviates from that at distances not too far from the variant site. The bin size used to count interactions is 5 kb. B, Variants in the AEC, aortic smooth muscle cell (ASMC) and macrophage–THP-1 (mTHP-1)-lipopolysaccharide (LPS) merged genome-wide association studies (GWAS)-Prom (GP) set and their interaction preferences with genes at distance 0 (no gene-jumping), 1 (nearest gene is jumped over), etc. Distal regions in the corresponding PD set are shown for reference. The GP interaction set is further split in equal-sized halves depending on the variants’ distance to its nearest gene (GP without and with a nearby gene, respectively). C, The coronary artery disease (CAD)–related variant rs12239436 (red box) interacts with 62 kb distant gene USP24 (gray box) in AEC (dotted line), ASMC (dashed,) and mTHP-1–LPS (solid). Strengths of interactions are represented with the P value recorded and indicated by arrow thickness. Our result set further include strong interactions with the 1.3 Mb distant previously CAD associated gene PLPP3 (also known as PPAP2B). Less significant interactions with nearby FYB2 (ASMC) and PRKAA2 (mTHP-1–LPS) are potentially bystanders. D, Earlier reported interaction between variant rs934937 and the EDN1 gene is, to a varying degree, present in both AEC patients. Not so in less relevant tissues ASMC and mTHP-1–LPS. E, Comparison of overlaps between PD dataset (AEC) vs 100 matched random datasets and CVD_GWAS and matched SNP datasets (see methods) shows that there is an enrichment for variants in linkage disequilibrium (LD) with CVD_GWAS found in PD dataset when the genomic distance between SNP and its LD proxy is ≤80 kb. No such enrichment was seen for random PD datasets vs real or random SNP sets. chr indicates chromosome; CVD, cardiovascular disease; SMC, smooth muscle cells; SNP, single nucleotide polymorphism; and SP, supporting pair.
