FIG 2.
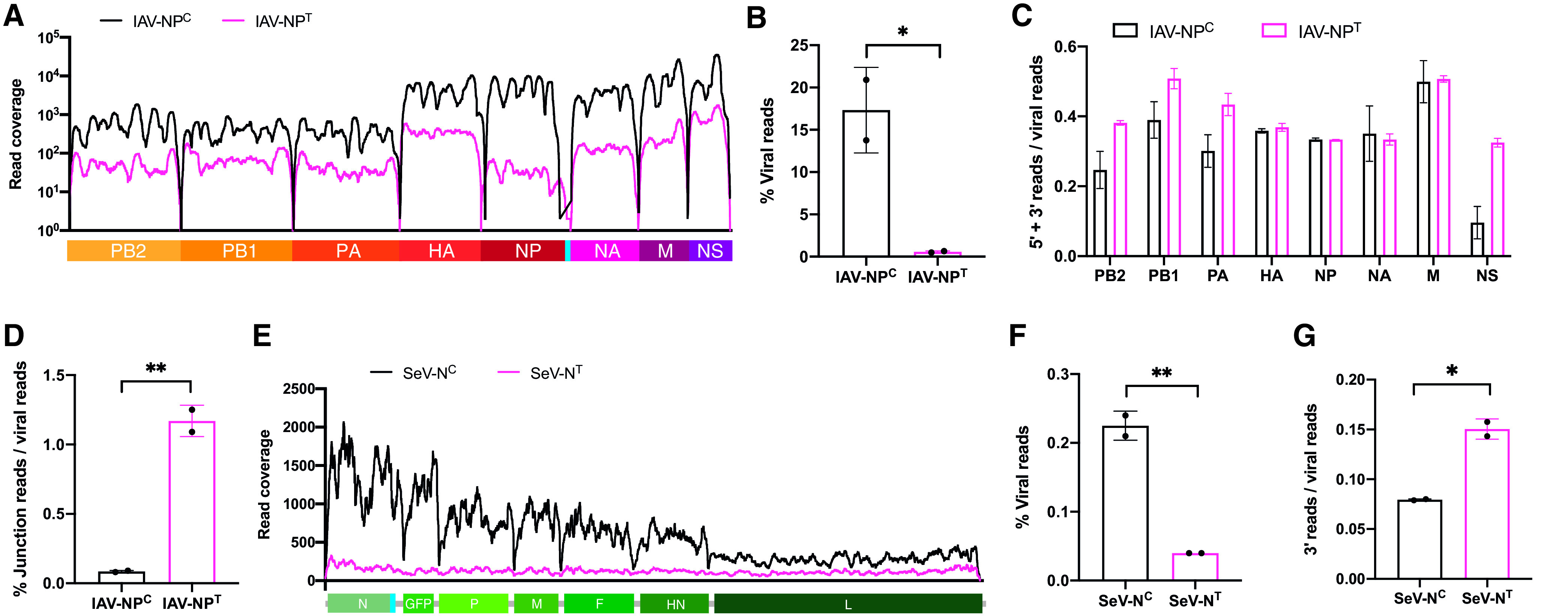
Limiting NP expression induces the production of defective viral genomes. (A) Read coverage along the IAV-NPC (black) or IAV-NPT (magenta) viral genomes in A549 cells infected at an MOI of 5 for 9 h after bulk mRNA-seq. (B) Mean percentage of viral reads over total mapped reads of total rRNA-depleted RNA-seq of IAV-NPC- or IAV-NPT-infected A549 cells (MOI of 5; 9 hpi). (C) Proportion of viral reads corresponding to panel A that map to the 5′ (15%)- and 3′ (15%)-terminal regions of each IAV segment. (D) Proportion of viral reads corresponding to panel B that map to noncanonical junction reads. (E) Read coverage along the SeV-NC (black) or SeV-NT (magenta) antigenome in A549 cells infected at an MOI of 5 for 9 h after ribosome-depleted total RNA-seq. (F) Mean percentage of viral reads over total mapped reads. (G) Proportion of viral reads that map to the 3′-terminal (15%) region of the SeV anti-genome. Data are derived from two independent biological replicates, with error bars depicting the standard deviation. Statistical significance was determined by an unpaired two-sample two-tailed t test. *, P < 0.05; **, P < 0.01.
