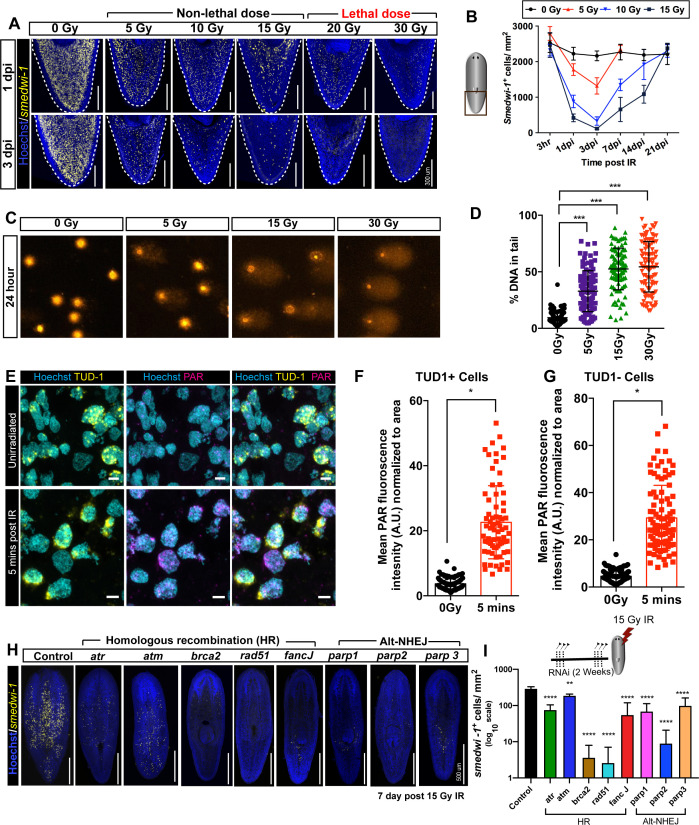
Figure 1. Planarian stem cell resistance to doses up to 15 Gy of gamma IR requires conserved DDR pathways.
(A) smedwi-1 FISH of planarians exposed to different doses of gamma IR (5, 10, 15, 20, and 30 Gy) after 1 and 3 days post-IR (dpi) showing a dose-dependent decrease in stem cell number. Scale bar: 300 μm. (B) Quantification of smedwi-1+ cells/mm2 (yellow) showing the repopulation kinetics of surviving stem cells after different doses of IR post-IR (n = 5 per dose, per time point). Results are expressed as mean ± SD. (C) COMET assay showing the extent of DNA breaks (comet shape) in isolated planarian cells at 24 hr after exposure to 5, 15, and 30 Gy of IR. (D) Quantification of the percentage of tail DNA in COMET assay post-IR at 24 hr. Results are expressed as mean ± SD. Each dot represents the tail DNA in individual planarian cells. (***p<0.0001, one-way ANOVA using Tukey’s multiple comparison test). (E) Double immunostaining with Anti-TUD-1 (Yellow) and Anti-PAR (Magenta) showing DNA damage in stem cells (Tud-1+) and post-mitotic differentiated cells (Tud-1−) at 5 min post 5 Gy IR. Nucleus is stained with Hoechst (blue). (F–G) Quantification of PAR fluorescence in Tud1+ and Tud1− cells normalised to the nuclear area in irradiated and unirradiated cells (0 Gy) (*p<0.0001. Student’s t-test). (H) Representative FISH showing stem cell repopulation in Control (gfp) RNAi and after knockdown of different DNA repair genes (involved in homologous recombination [atr, atm, brca2, fancJ, rad51] and Alt-NHEJ [parp1, parp2, parp3]) after 7 days post 15 Gy IR. Gene name represents the RNAi condition. (I) Repopulation of smedwi-1+ cells/mm2 in DDR RNAi worms after 7 days post 15 Gy IR (n = 5 per condition). Results are expressed as mean ± SD in log10 scale (***p<0.0001, **p<0.001, one-way ANOVA using Tukey’s multiple comparison test).