Figure 1.
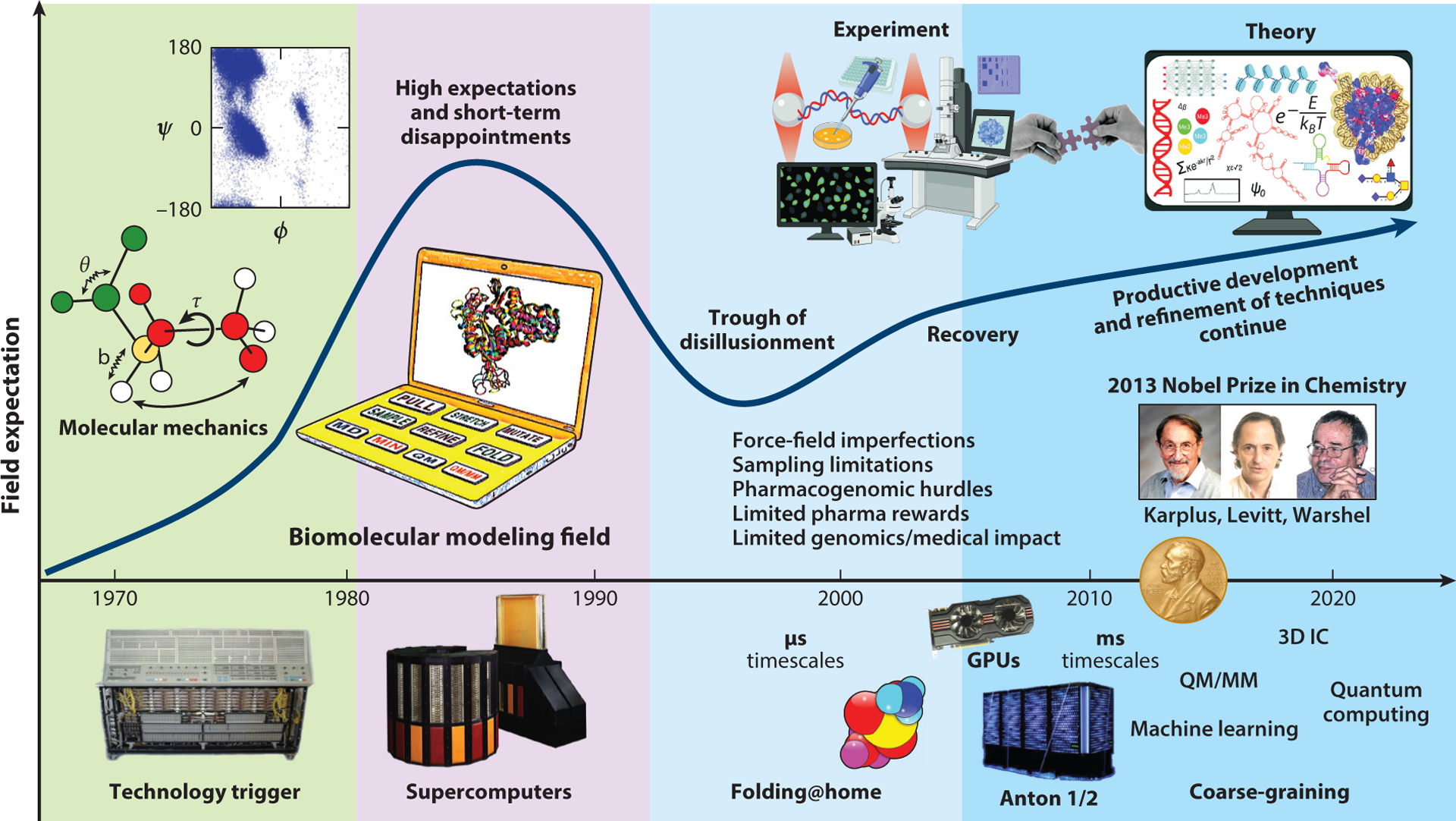
Expectation curve for the field of biomolecular modeling and simulation. The field started with comprehensive molecular mechanics efforts, and it took off with the increasing availability of fast workstations and, later, supercomputers. Following unrealistically high short-term expectations and disappointments concerning the limited medical impact of modeling and genomic research on human disease treatment, better collaborations between theory and experiment have ushered the field to its current productive stage. Problems realized in the decade between 2000 and 2010 and later addressed include force field imperfections, conformational sampling limitations, some pharmacogenomics hurdles, and limited medical impact of genomics-based therapeutics for human diseases. Technological innovations that have helped drive the field include distributed computations and the advent of the use of graphic processing units for biomolecular computations. The molecular-dynamics-specialized supercomputer Anton made it possible in 2009 to reach the millisecond timescale for explicit-solvent all-atom simulations. The 2013 Nobel Prize in Chemistry awarded to Levitt, Karplus, and Warshel helped validate a field that lagged behind experiment and propel its trajectory. Abbreviation: QM/MM, quantum mechanics/molecular mechanics.
