Figure 3.
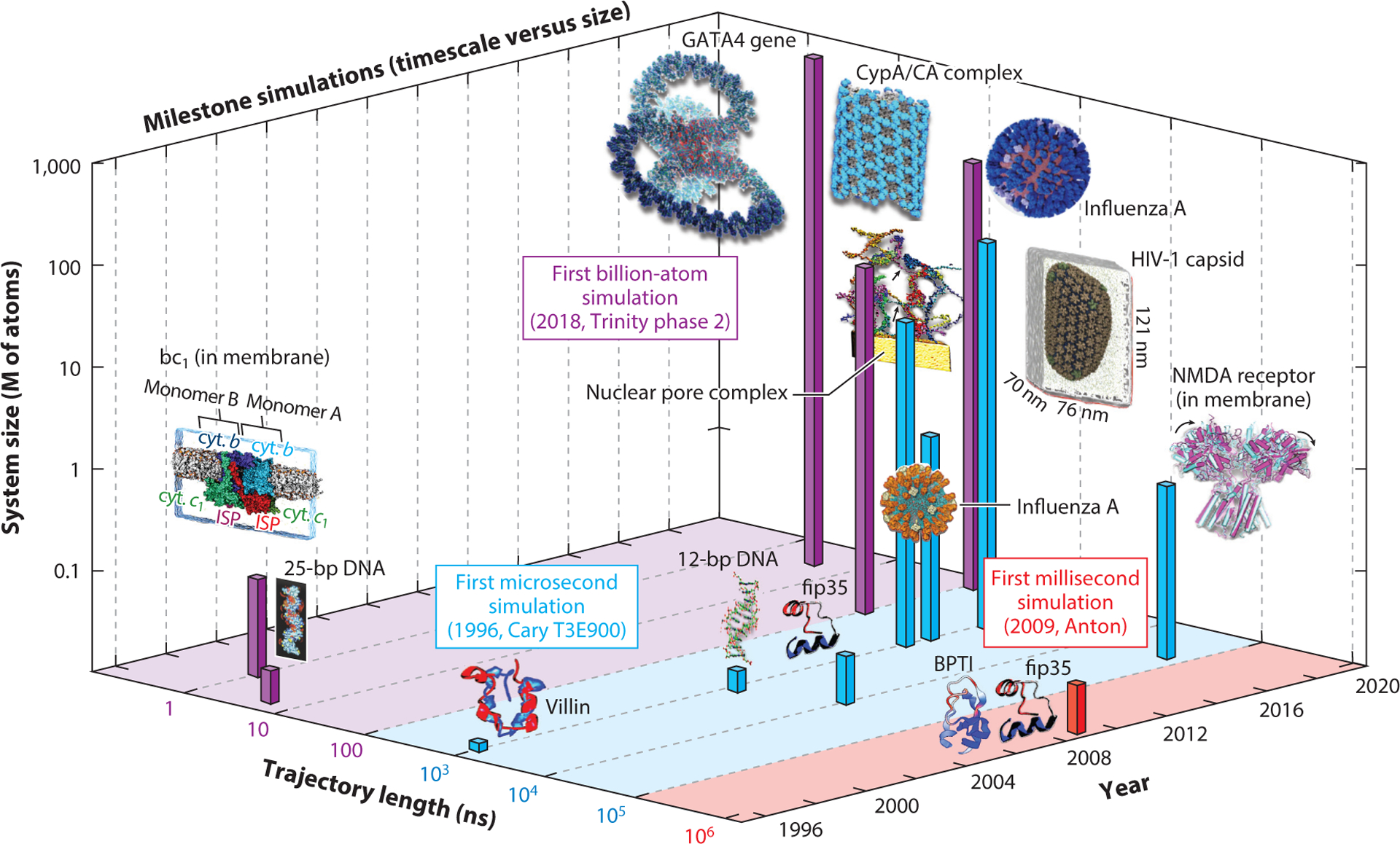
Milestone simulations in biomolecular modeling showing evolution in molecular dynamics timescale and system size. Consistent with Figure 2f, we assume that computations were performed one year before publication, except for the publications in 1998, for which the calculations were performed in 1996. The simulated systems in temporal order are: 25-base-pair DNA (5 ns and ~21k atoms) (204), villin protein (1 μs and 12k atoms) (43), bc1 membrane complex (1 ns and ~91k atoms) (70), B-DNA dodecamer (1.2 μs and ~16k atoms) (141), Fip35 protein (10 μs and ~30k atoms) (49), Fip35 and BPTI proteins (100 μs for Flip35 and 1 ms for BPTI, and ~ 13k atoms) (182), nuclear pore complex (1 μs and 15.5M atoms) (50), influenza A virus (1 μs and >1M atoms) (158), NMDA receptor in membrane (60 μs and ~507k atoms) (187), tubular CypA/CA complexes (100 ns and 25.6M atoms) (111), HIV-1 fully solvated empty capsid (1 μs and 64M atoms) (143), GATA4 gene (1 ns and 1B atoms) (83), and influenza A virus H1N1 (121 ns and ~160M atoms) (44).
