Figure 4.
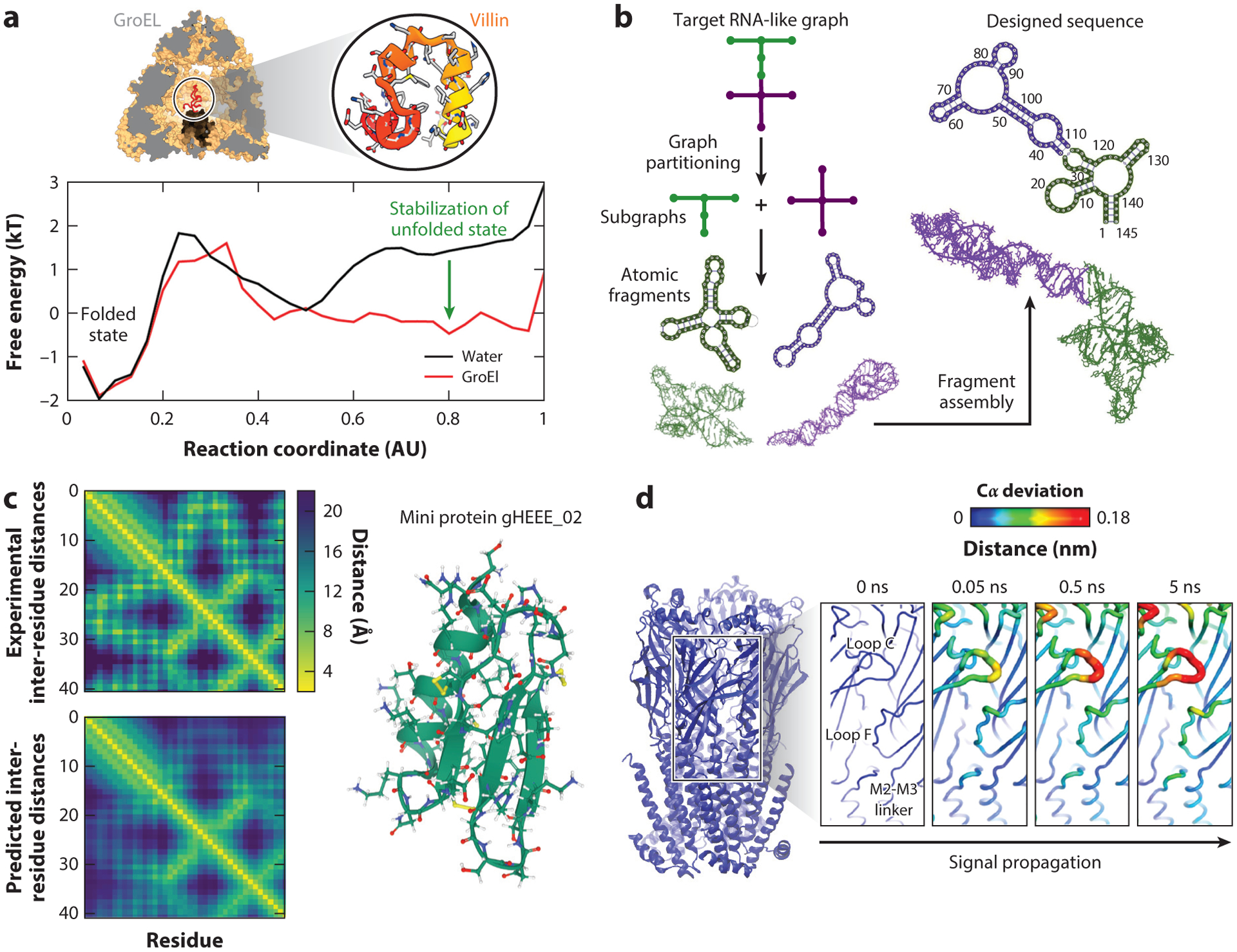
Examples of modeling successes. (a) Protein folding. The folding of a small protein (top right), villin, inside the GroEL protein cavity (top left) is compared to its folding in bulk. The corresponding energy profiles of folding show significant differences in the shape of the unfolded basin, indicating the role of the chaperonin protein in stabilizing unfolded states. Panel adapted with permission from Reference 148. (b) RNA novel motif design. Shown is a flowchart of the computational pipeline for design of RNA-like tree graph topologies (73, 75). In these tree graph representations of RNA secondary structures, edges denote stems, and vertices denote loops, bulges, and junctions (169). We use graph partitioning to segment the target RNA-like graph into subgraphs, extract the corresponding atomic fragments from our RAG-3D database, construct a new sequence or structure using fragment assembly, and screen the top-scoring sequences using RNA 2D structure prediction programs to produce successful sequences that will fold onto the target RNA-like topology (73–75, 117). (c) AlphaFold performance on prediction of inter-residue distances. Inter-residue distance distributions are obtained from the experimental structure (top) and predicted structure (bottom) of the miniprotein gHEEE02 (right), showing good agreement. Distance maps were obtained from https://deepmind.com. (d) Cloud computing to accelerate molecular dynamics. Extensive nonequilibrium simulations of nicotine unbinding from the nicotinic acetylcholine receptor are shown. The Cα fluctuation levels (colored on a scale from blue to red as fluctuations increase) show the sequence of structural changes coupled to the unbinding of the nicotine from the receptor that leads to its activation through a conformational change. Panel adapted with permission from Reference 135.
