Figure 5.
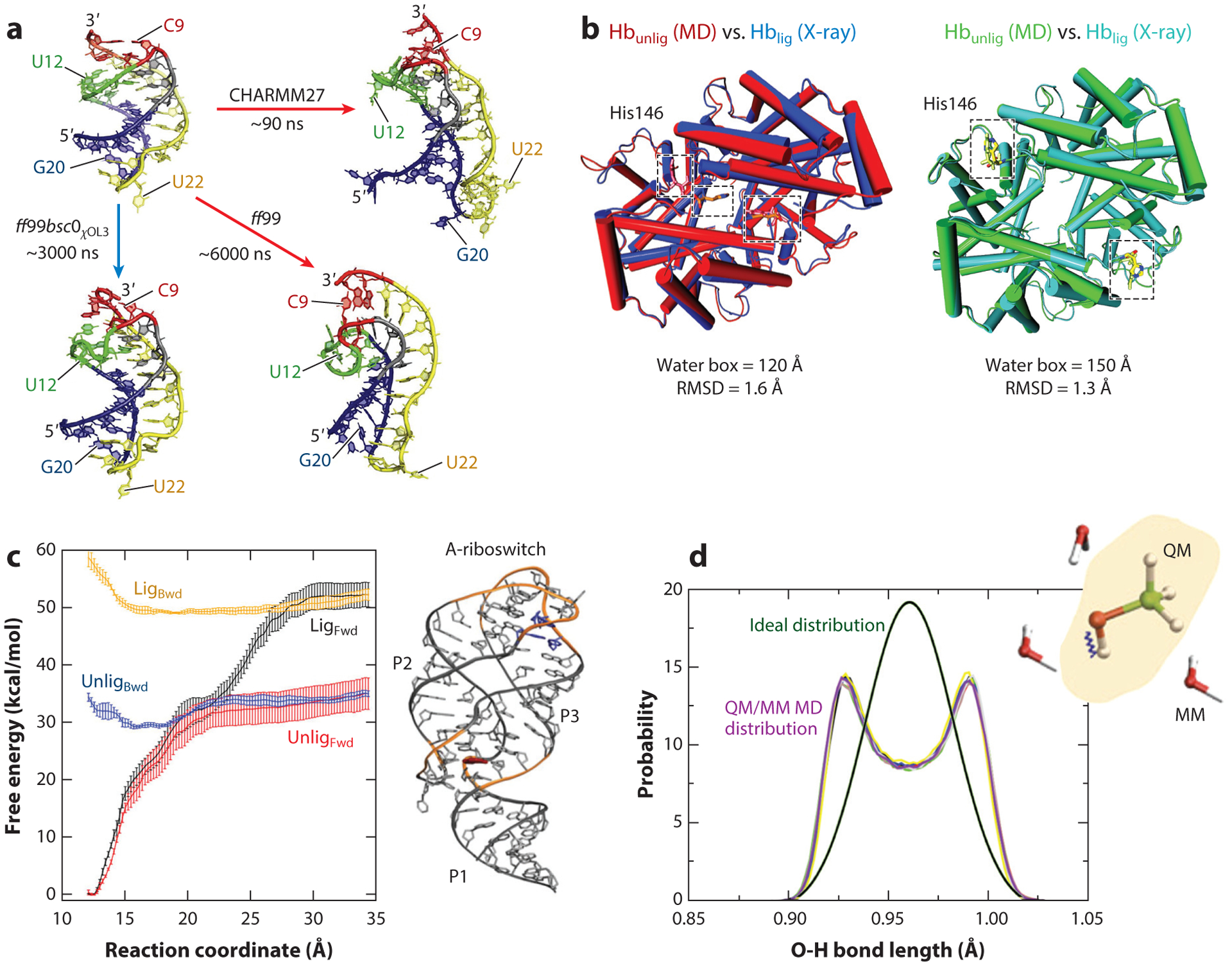
Examples of modeling failures. (a) RNA force fields. Riboswitch aptamer simulations with different force fields indicate that structural details and stabilities are force field dependent. With the CHARMM and ff99 force fields, the aptamer pseudoknotted fold is distorted compared to the X-ray structure, whereas with the ff99bsc0χOL3 force field, the aptamer fold is maintained. Panel adapted with permission from Reference 13. (b) Configurational sampling. A large water box is required to stabilize the unliganded tetramer of the hemoglobin. The unliganded structure obtained with molecular dynamics simulations using a water box of 150Å aligns well with the corresponding experimental structure (right), whereas the unliganded structure obtained with a water box of 120Å is instead similar to the experimental structure of the liganded conformation (left). Panel adapted with permission from Reference 45. (c) Advanced sampling techniques. Different free energy profiles are obtained with umbrella sampling for the forward (Fwd) and backward (Bwd) processes of a conformational change for a riboswitch in the presence (Lig) and absence (Unlig) of a ligand, a consequence of poor convergence of the simulations. Panel adapted with permission from Reference 38. (d) QM/MM simulations. Simulations of solutes treated at the quantum-mechanical level embedded in a rigid water model treated by classical molecular mechanics can lead to poor sampling due to insufficient coupling between the two regions. Shown is the incorrect distribution of the methanol O-H bond length obtained from QM/MM simulations (violet curve) compared to the ideal distribution (green curve). Panel adapted with permission from Reference 65. Abbreviations: MD, molecular dynamics; QM/MM, quantum mechanics/molecular mechanics.
