Figure 6.
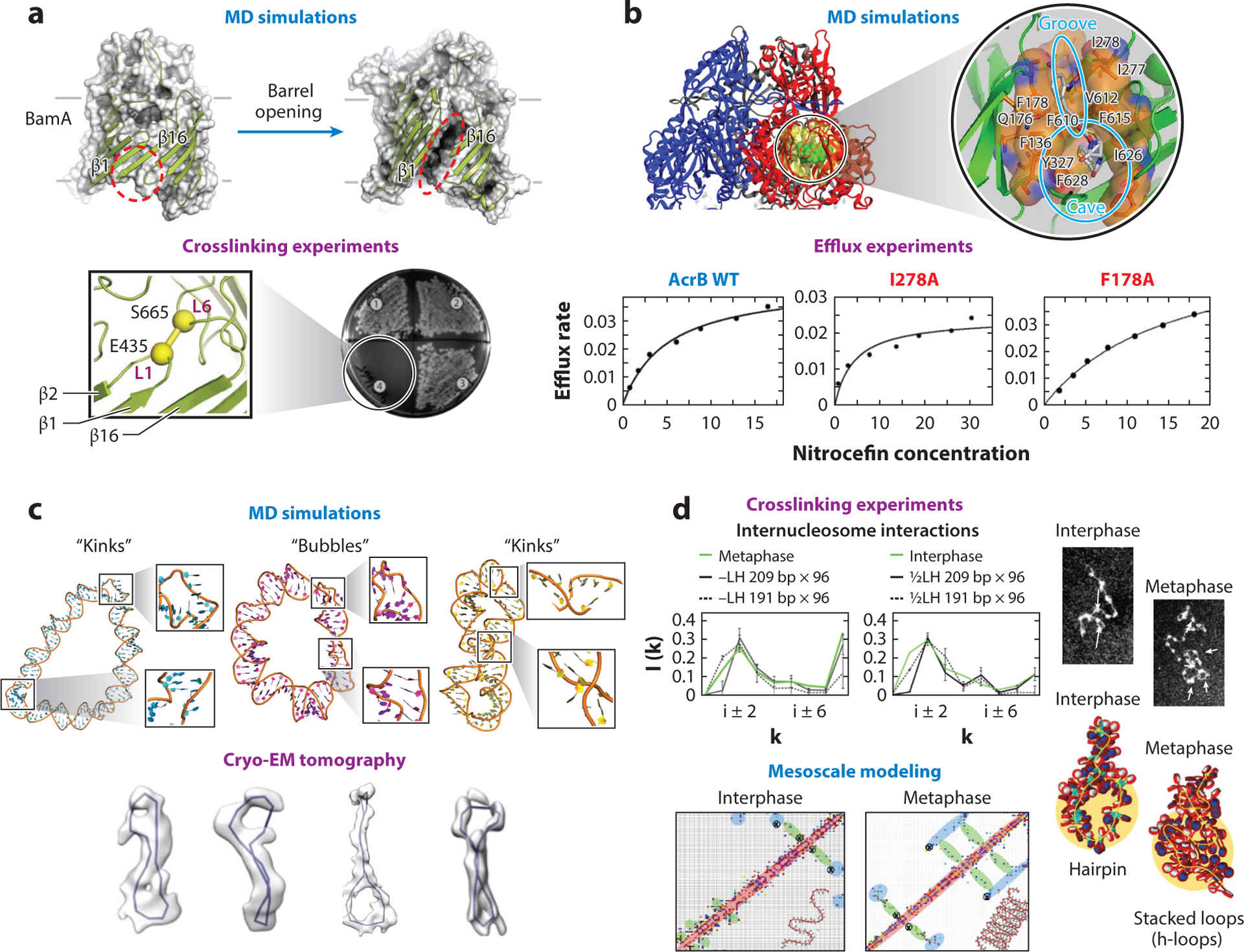
Interplay modeling experiment. (a) Molecular dynamics (MD) simulations of the bacterial protein BamA predicted a membrane insertion mechanism by lateral opening of the β barrel that involves the strands β1 and β16 (top). Crosslinking experiments created artificial disulfide bonds between loops L1 and L6 that connect both β strands and confirmed that BamA function was inhibited (bottom). Panel adapted with permission from Reference 133. (b) MD simulations of the AcrB multidrug transporter with the drug nitrocefin predicted a drug-binding pocket that includes residues such as I278 and F178 (top). Mutagenesis and biophysics experiments, which measured the efflux rate at different nitrocefin concentrations, confirmed the role of these residues in the binding of the drugs (bottom). Panel adapted with permission from References 194 and 89. (c) MD simulations of DNA minicircles predicted the formation of bubbles and kinks under torsional stress (top). Electron cryo-tomography experiments confirmed the formation of such geometries in DNA minicircles (bottom). Panel adapted with permission from Reference 69. (d) Chromatin crosslinking experiments show increased long-range internucleosome contacts without loss of zigzag short-range contacts for metaphase chromatin, compared to interphase chromatin (top; interaction patterns in green). Mesoscale model of fibers typical of interphase [0.5 LH/nucleosome and nucleosome repeat length (NRL) = 191 bp] and metaphase (no LH and NRL = 209 bp) chromatin provide the folding mechanism of hierarchical looping (or stacked loops in 3D) to explain such increases in long-range contacts with maintenance of short-range contacts. This can be observed in the computed contact maps and fiber structures (bottom), and in the interaction patterns for fibers with NRL = 191 or 209 bp (top, black solid and dashed lines) without LH (left) and with 0.5 LH/nulceosome (right) (56).
