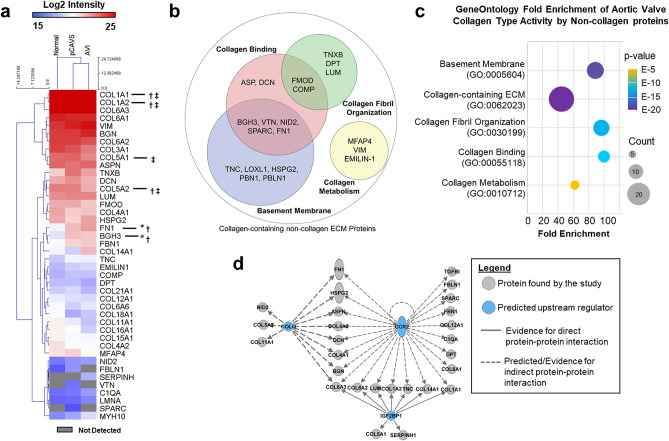
Figure 5.
Extracellular matrix proteomics implicates collagen binding and collagen organization pathways. (A) Aortic valve extracellular matrix proteins identified by the collagen-targeting proteomics. A total of 18 collagen proteins were identified with COL1A1, COL1A2 and COL6A3 being the most abundant collagens. Protein level quantification, which includes all peptides both unmodified and modified, revealed significant differences based on valve diagnoses in COL1A1, COL1A2, COL5A1, BGH3, FN1, and COL5A2. *Mann–Whitney U adj. p-value < 0.05 normal AV compared to pCAVS, †Mann–Whitney U adj. p-value < 0.05 normal AV compared to AVI, ‡Mann–Whitney U adj. p-value < 0.05 AVI compared to pCAVS. (B) Gene Ontology Functional classification of non-collagen type proteins identified in the proteomics dataset. (C) Gene Ontology Fold Enrichment analysis of collagen type function by non-collagenous proteins identified by proteomics, Fisher’s exactp-value ≤ 1.84E–5, false discover rate ≤ 6.66E–3. Proteins associated with collagen binding had the highest degree of fold enrichment, followed by collagen fibril organization. (D) Upstream analysis of the ECM proteome by Ingenuity Pathways Analysis to identify potential regulators that explain directional changes in aortic valve ECM proteins. Primary regulators affecting the proteome were C–C Motif Chemokine Receptor 2 (CCR2; Fisher’s Exactp-value 1.13E–39), Collagen Like Tail Subunit Of Asymmetric Acetylcholinesterase (COLQ; Fisher’s Exactp-value 9.67E-19), (Insulin Like Growth Factor 2 MRNA Binding Protein 1 (IGF2BP1, Fisher’s Exactp-value 1.05E–18). Normal n = 4, pCAVS n = 11.