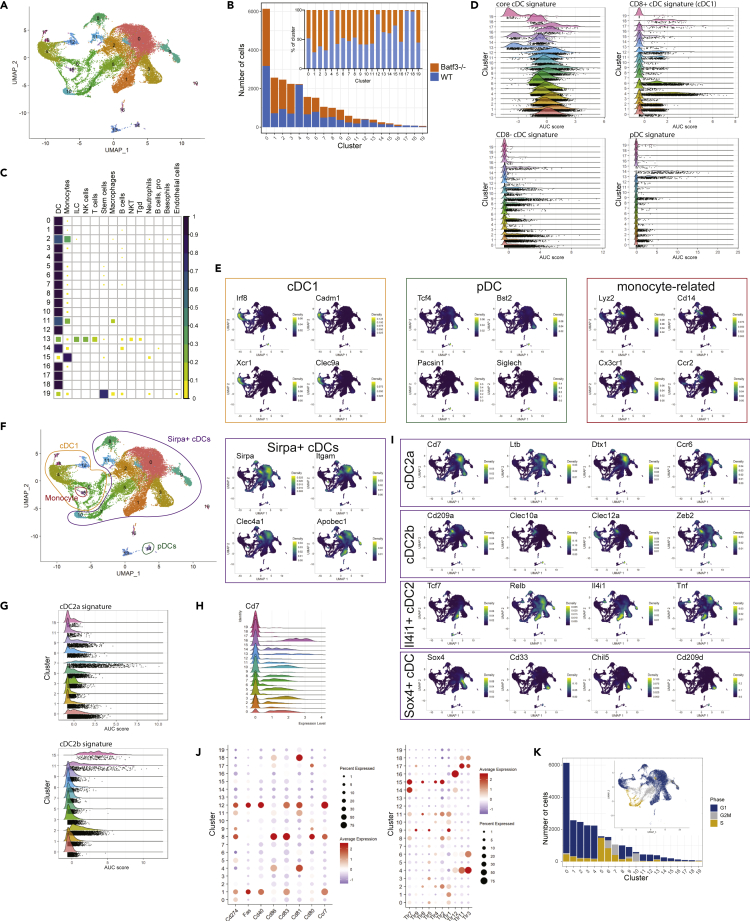
Figure 2.
Splenic cDC compartment consists of heterogeneous cell states of cDC1 and cDC2
Sorted MHCII+ CD11c+ splenic DCs of C57BL/6 and Batf3−/− mice (see Figure S2) were incubated with Total-Seq antibodies and sequenced using the 10X Genomics droplet-based sequencing platform. Application of the Seurat pipeline resulted in 20 clusters at resolution 0.5.
(A) UMAP depicting clusters based on Louvain algorithm using the FindNeighbors function in Seurat.
(B) Proportion and number of cells assigned to WT and Batf3−/− genotypes.
(C) SingleR prediction of clusters using the ImmGen database-identified DC clusters and non-DC contaminations.
(D) Gene set enrichment analysis (GSEA) using DC gene signatures from (Miller et al., 2012) and the AUCell package (see Table S2).
(E) Density plots of key canonical DC and monocyte-delineating features using the Nebulosa package (Jose Alquicira-Hernandez, 2020).
(F) Definition of cluster groups for cDC1, Sirpa+ DCs, pDCs, and monocytes.
(G) GSEA of Sirpa+ DC clusters and monocytes (C15) using cDC2a and cDC2b gene signatures from Brown et al. (2019) and the AUCell package (see Table S2).
(H) Ridgeplot of Cd7 expression.
(I) Density plots of key genes delineating Sirpa+ cDC clusters using the Nebulosa package.
(J) Expression of maturation-associated and TLR genes.
(K) Distribution and proportion of cells assigned to cell cycle phases based on the expression of gene signatures of the G1, S, and G2/M phases (see Table S5).