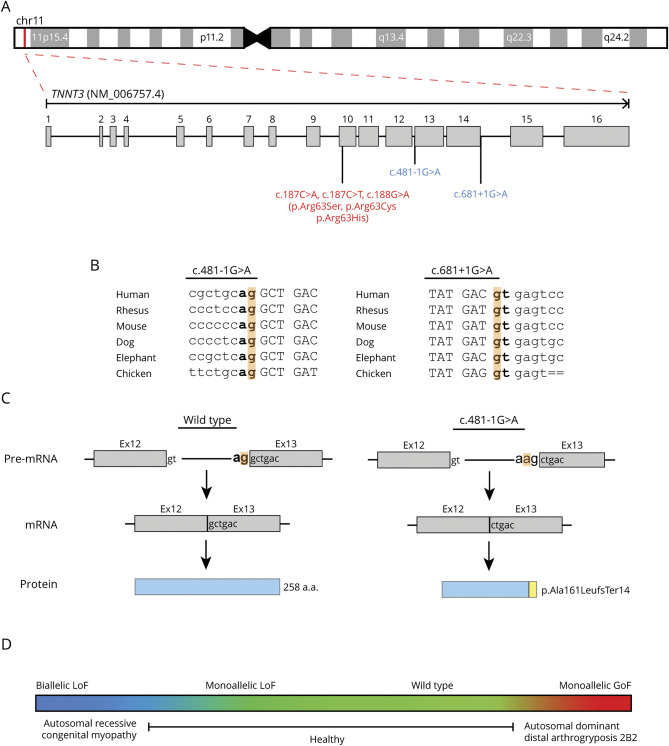
Figure 2. TNNT3 Variant Details and Model of TNNT3-Related Disease.
(A) Exon structure of TNNT3 (NM_006757.4). TNNT3 is located on chromosome 11p15.5. Variants linked to autosomal recessive congenital myopathy are indicated in red, whereas variants associated with autosomal dominant DA2B2 are in blue. (B) Location and conservation of TNNT3 splice variants c.481-1G>A and c.681+1G>A. Intronic sequences are indicated in lowercase. Exonic sequences are indicated in upper case. Consensus splice sites are indicated in bold. The variant location in highlighted in orange. (C) Predicted impact of c.481-1G>A on splicing and final protein product. Consensus splice sites are indicated in bold. The variant location is highlighted in orange. Normal splicing and translation of wild-type TNNT3 mRNA (NM_006757.4) results in a 258 amino acid protein. The c.481-1G>A substitution is predicted to cause loss of the consensus splice acceptor and result in the creation of a new splice acceptor shifted +1 bp in the 3′ direction. This new splice acceptor should result in a −1 frameshift and therefore is predicted to cause a prematurely truncated protein, p.Ala161LeufsTer14. (D) TNNT3 disease model. Genotype is indicated above the colored box, and phenotype is displayed below. Monoallelic LoF TNNT3 variants are tolerated (green), whereas biallelic LoF TNNT3 variants (blue) cause severe congenital myopathy with respiratory and bulbar muscle weakness and scoliosis. In contrast, gain-of-function missense variants increasing contractility (red) result in autosomal dominant DA2B2.