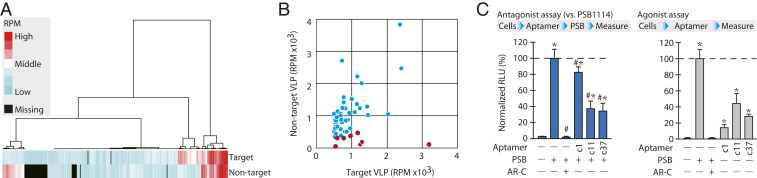
Fig. 2.
Selection of candidate aptamers from HTS data. (A) Heatmap analysis. The difference in the read number (RPM) of each aptamer, composing the enriched library to VLPs expressing target GPCR (P2RY2) and nontarget GPCR (EDNRB), was visualized by a heatmap hierarchical clustering. (B) Scatter plot from HTS data. Based on the RPM of the data from sequences bound to the target VLP (x-axis) and that from sequences bound to the nontarget VLP (y-axis), 51 enriched sequences representing each cluster were shown. The eight sequences that were enriched more than twofold in target-associated sequences compared to nontarget-associated sequences are shown in red. Each dot represents the sequences showing the highest RPM in each cluster. (C) Effects of aptamers on the P2RY2 activation. Inhibitory potency of the aptamers (c1, c11, and c37, final 2.5 μM) and chemical antagonist AR-C118925XX (AR-C, final 10 μM) were examined when the cells overexpressing P2RY2 were posttreated by chemical agonist PSB1114 (PSB, final 100 nM) (Left). In the agonist assay (Right), the cells overexpressing P2RY2 were treated by aptamers (final 5 μM) and PSB (final 100 nM). The pretreatment of AR-C at a concentration of 10 μM was performed to confirm the PSB specificity. Data represent the mean ± SD (n = 3 independent experiments). The values were expressed as relative luminescent units (RLU shown in %) to the 100 nM PSB1114 level without aptamer after subtraction of basal LU in control cells without treatment. #P < 0.05 versus PSB1114 treatment group without aptamer in the antagonist assay; and *P < 0.05 versus no treatment group in both assays.