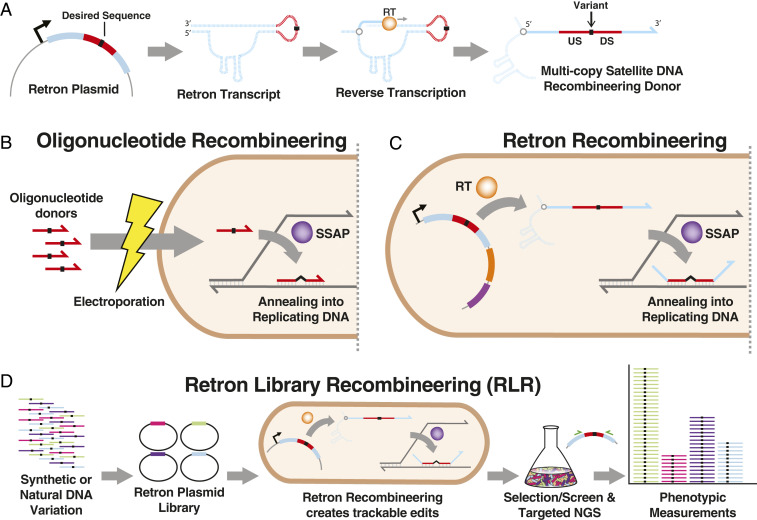
Fig. 1.
Overview of retron recombineering. (A) The retron msr/msd region undergoes transcription, then targeted reverse transcription catalyzed by retron reverse transciptase (RT), producing multicopy satellite DNA. When a sequence (red) containing homology upstream (US) and downstream (DS) of a sequence alteration (black) is introduced, this DNA functions as a recombineering donor. Dashed lines depict RNA, and solid lines depict DNA. (B) In oligonucleotide recombineering, synthetic oligonucleotide donors introduced into bacteria anneal to replicating DNA, directed by a single-stranded annealing protein (SSAP). This introduces desired sequence alterations (black) into the genome. (C) Retron recombineering uses RT-derived DNA as a donor, rather than transformed oligonucleotides, but similarly incorporates these into replicating DNA using an SSAP. (D) Libraries of synthetic or natural DNA variants can be incorporated into retrons to perform recombineering. This produces mutant cells bearing a retron plasmid, available for targeted amplicon sequencing using complementary primers (green) to measure mutant abundance in multiplex.