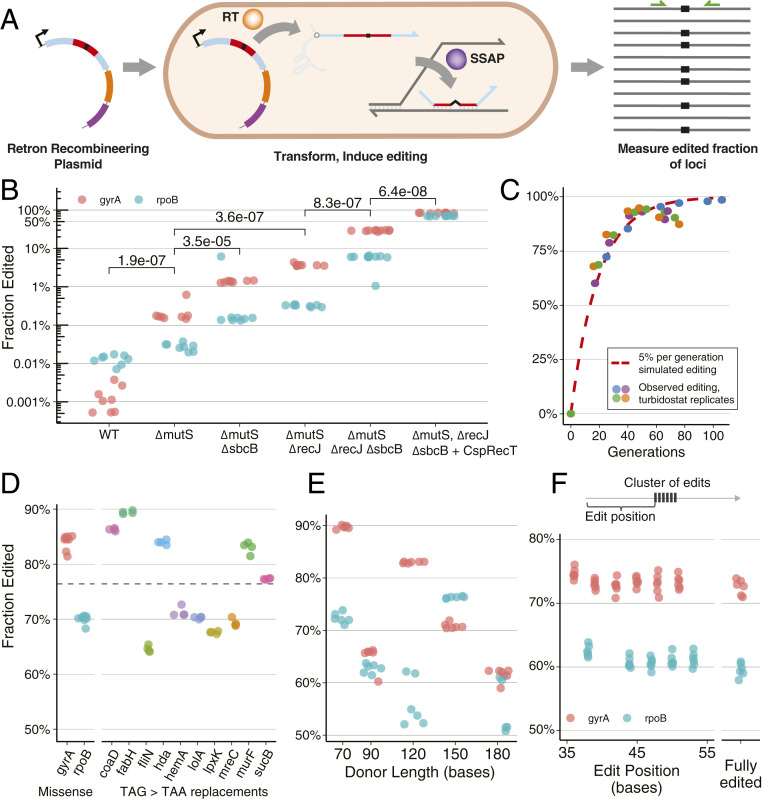
Fig. 2.
Characterization and optimization of retron recombineering. (A) A retron plasmid creates multicopy ssDNA via a retron RT. This DNA is incorporated into the genome via a coexpressed SSAP. Editing is observed by amplicon NGS, facilitated by primers targeting the locus (green). (B) Optimization of RLR. Colors differentiate gyrA and rpoB edits. Biological replicates are indicated with dots. Two-sided, unpaired parametric t tests were performed between genotypes indicated with brackets, and P values are displayed. (C) Editing of cultures in a turbidostat, with continuous growth and induction. mutS sbcB recJ genotype was used, transformed with gyrA-editing plasmid, expressing Red as an SSAP. These data are shown alongside a simulated editing trajectory of an allele with neutral fitness effect, editing at 5% per generation. (D) Edited fraction observed across alleles, after approximately 20 generations of induction and batch growth. A retron plasmid containing CspRecT as an SSAP was expressed in mutS recJ sbcB in all cases. Results of individual replicates are shown as dots. Results for gyrA and rpoB missense alleles (in B) are shown again for comparison alongside TAG > TAA stop codon editing for 10 essential genes. The mean edited fraction achieved across the 12 loci, 76.4%, is indicated by the dashed line. (E) The effect of retron length on editing. Six replicates for each experiment are indicated with dots colored by locus. (F) Integration of a cluster of mismatched bases in a retron donor. Editing of each position across eight replicates is shown for gyrA and rpoB alleles. Observations having all mutations are reported as fully edited.