Figure 3. Enzymes and SBP for HMO utilization by infant gut-associated bifidobacteria.
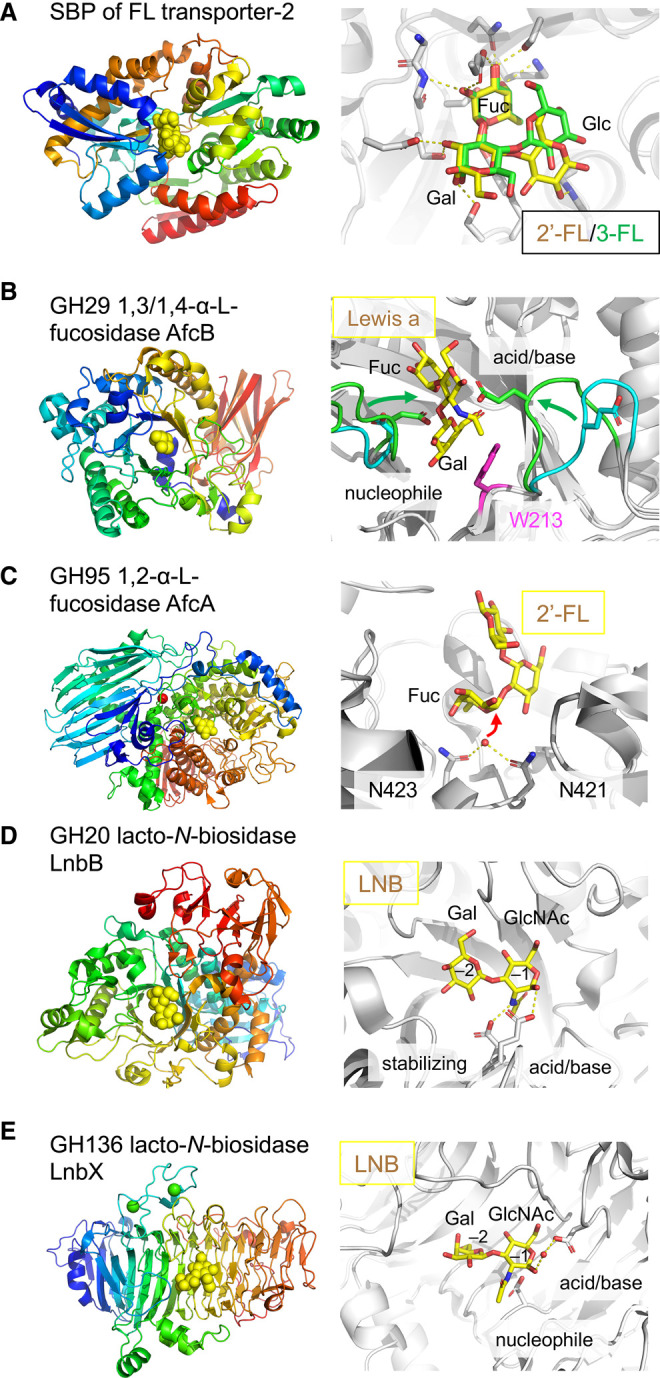
(A) Fucosyllactose binding protein (SBP) of the FL transporter-2 from B. infantis JCM 1222. Left panel, overall structure (PDB: 6HUR) shown with 2′-fucosyllactose (2′-FL) as yellow spheres. Right panel, the substrate-binding site is shown as a superimposition of the complex structures with 2′-fucosyllactose (2′-FL, yellow, PDB: 6HUR) and 3-fucosyllactose (3-FL, green, PDB: 6HUS). (B) GH29 1,3/1,4-α-L-fucosidase AfcB from B. infantis ATCC 15697. Left panel, overall structure (PDB: 3UET) is shown with fucose as yellow spheres. Right panel, the induced-fit motion of the active site loops shown as a superimposition of apo (cyan, PDB: 3MO4) and complex (green, PDB: 3UET) structures with Lewis a trisaccharide (yellow sticks). The side chains of the catalytic residues (green sticks) were modelled from the D172A/E217A double mutant structure. The aromatic stacking platform for Gal (Trp213, magenta) is also shown. (C) GH95 1,2-α-L-fucosidase AfcA from B. bifidum JCM 1254. Left panel, overall structure (PDB: 2EAC) is shown with deoxyfuconojirimycin as yellow spheres. Right panel, the active site is shown as a composite of the 2′-FL complex (PDB: 2EAD) and the wild-type enzyme (PDB: 2EAC). 2′-FL, two catalytic asparagine residues, and the nucleophilic water are shown as yellow sticks, white sticks, and a red sphere, respectively. (D) GH20 lacto-N-biosidase LnbB from B. bifidum JCM 1254. Left panel, overall structure (PDB: 4H04) is shown with LNB as yellow spheres. Right side, the active site showing LNB. (E) GH136 lacto-N-biosidase LnbX from B. longum JCM 1217. Left panel, overall structure (PDB: 5GQF) is shown with LNB as yellow spheres. Right side, the active site showing LNB. The side chains of the catalytic residues are shown in (D) and (E).
