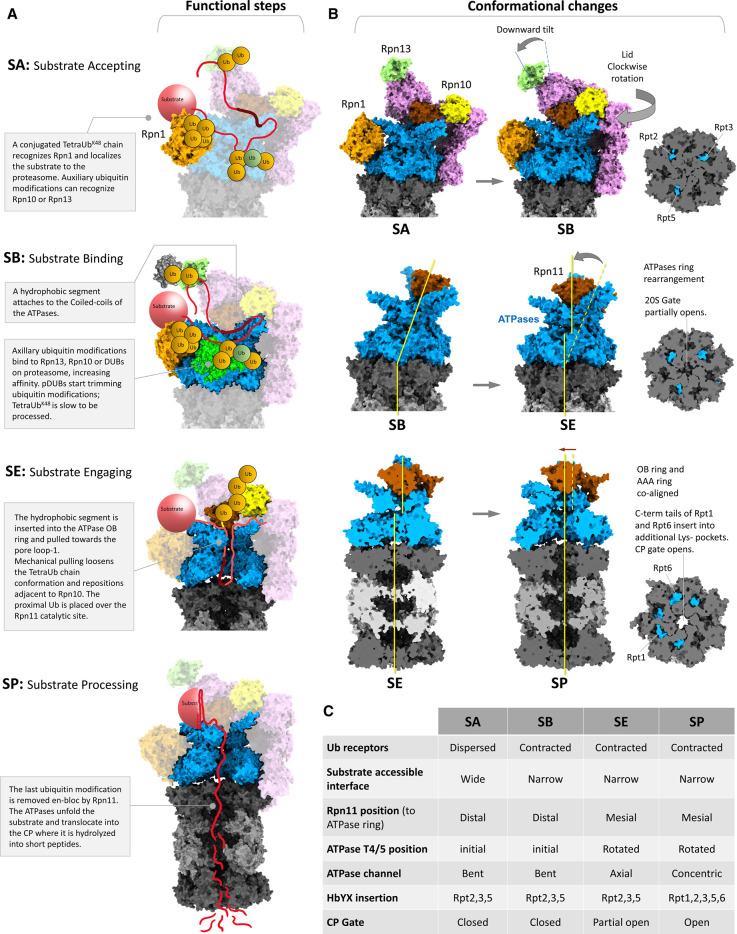
Figure 4. Functional steps and conformational changes during substrate processing by 26S proteasomes.
(A) We describe four steps during proteasome function: SA, substrate accepting; SB, substrate binding; SE, substrate engagement and SP, substrate processing. The substrate model illustratively depicts a multiple ubiquitinated-conjugate modified by a Lysine48-linked tetraUb and a branched-mixed polyubiquitin chain. (B) Illustration of conformational changes during transition from one step to the next. Proteasome models are generated using different PDB structures of 26S proteasomes in various conformations (PDB: 6j2x, 6j30 and 6j2n). The three cross-sectional views of 20S CP near upper α-ring show; a three-tail insertion with close gate (SA), a three tail-insertion with partial open gate (SE) and a five-tail insertion with open gate (SP) are generated using PDB: 6j2x, 6j30 and 6msk, respectively. (C) A table summarising the major characteristic features of each functional step following substrate binding.