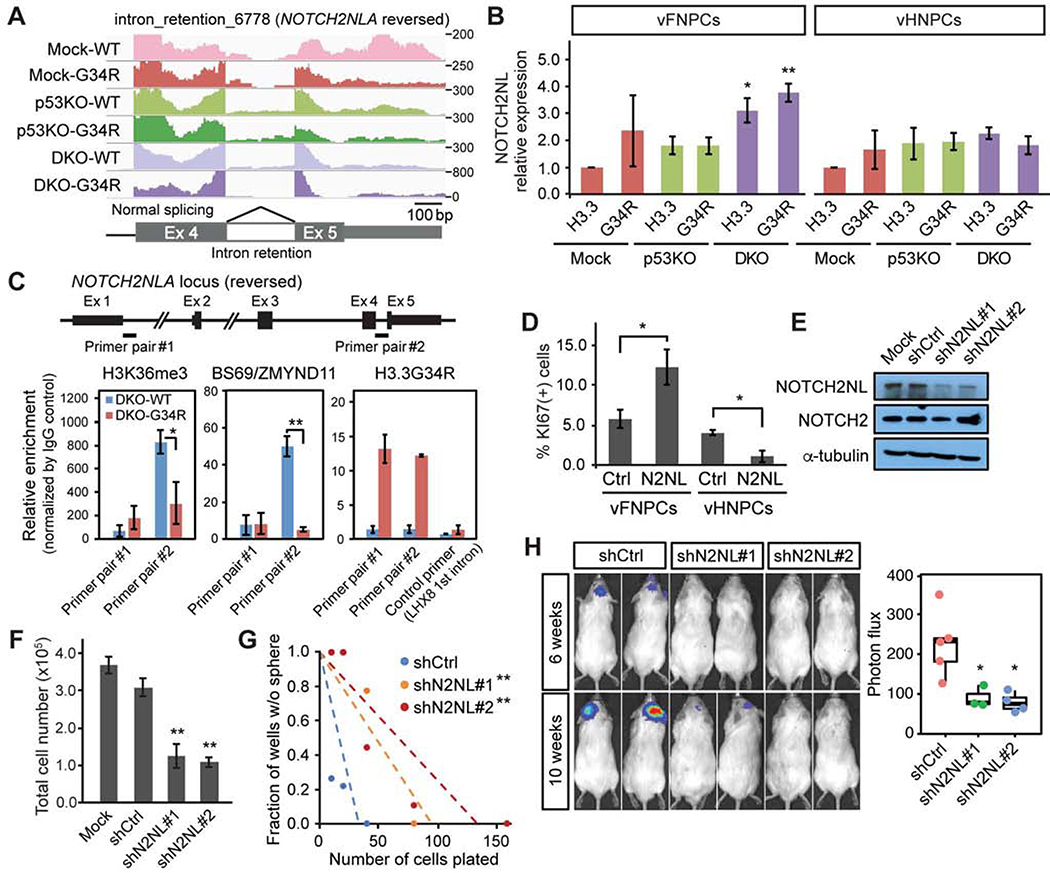
Figure 5. Upregulation of NOTCH2NL genes contributes to tumorigenicity in H3.3G34R-mutant tumors.
(A) RNA-seq read mapping at the last exons of NOTCH2NL gene shows decreased intron retention in triple-mutant vFNPCs (DKO-G34R).
(B) Expression of NOTCH2NL is increased by the mutations in vFNPCs, but not in vHNPCs. Bars indicate mean ± S.E.M. (n = 3–5).
(C) ChIP-PCR shows decreased H3K36me3 and BS69/ZMYND11 binding at the intron retention site of NOTCH2NL (primer pair #2), but not at the first intron (primer pair #1). Occupancy by H3.3G34R is confirmed by a specific antibody. A primer for the first intron of LHX8 was used as a control. Bars indicate mean ± S.E.M. (n = 3–4).
(D) Overexpression of NOTCH2NL (N2NL) enhances proliferation in p53KO vFNPCs, but not in p53KO vHNPCs. Bars indicate mean ± S.E.M. (n = 3–6).
(E) Western blotting confirms specific knockdown of NOTCH2NL by two independent small hairpin RNAs (shRNAs).
(F) Knockdown of NOTCH2NL in H3.3G34R-mutant patient-derived cell line (HSJD002) significantly reduces the total cell number. Bars indicate mean ± S.D. (n = 4).
(G) Limiting dilution assay shows a decrease in sphere-forming capacity by NOTCH2NL knockdown.
(H) Bioluminescence imaging shows suppressed in vivo growth of H3.3G34R-mutant cell line by NOTCH2NL knockdown (n = 4–5). P values were calculated by two-sided Welch’s t test (B, C, D, F, H) or the likelihood-ratio test (G). *P < 0.05, **P < 0.01.